Keywords
carbapenem resistance, carbapenemase enzymes, gram-negative bacilli, Modified Hodge Test, PCR, bla IMP gene, bla OXA48
carbapenem resistance, carbapenemase enzymes, gram-negative bacilli, Modified Hodge Test, PCR, bla IMP gene, bla OXA48
Carbapenems are beta-lactam antibiotics often used as last resort antibiotics for treating infections caused by multidrug resistant Gram-negative bacilli, since they have the broadest spectra among all beta-lactams1,2. carbapenemases (also known as carbapenem hydrolyzing enzymes) represent the most versatile family of beta-lactamases3. Beta-lactamases can be classified according to their functional and molecular properties. Functional classification divides beta-lactamases into four major functional groups (1, 2, 3, and 4), whereas in molecular classification Ambler and others have classified beta-lactamases into four classes (A–D) based on amino acid sequence. In addition, based on the active site of the enzyme, they classified beta-lactamases into serine-beta-lactamases (class A,C and D) and metallo-beta-lactamases (class B)3. https://www.microrao.com/micronotes/pg/carbapenemases.pdf
Oxacillinases (OXAs) (class D), so called because of their ability to hydrolyze Oxacillin, are classified according to their hydrolysis spectrum. Broad-spectrum OXA enzymes are able to hydrolyze carbapenems. This family is plasmid encoded and they have been identified mainly in Enterobacteriaceae and Pseudomonas aeruginosa. Currently, there are 239 OXA enzymes, of which at least 37 are carbapenemases and at least 9 are extended spectrum beta-lactamases. The first OXA enzyme with carbapenemase activity was detected in 1985 in an Acinetobacter baumanii isolate from Scotland, and OXA-48 cluster is the most important among class D carbapenemases3,4. https://www.microrao.com/micronotes/pg/carbapenemases.pdf
IMP (for “active on Imipenem”) a metallo-beta-lactamase (class B), was first detected in 1990 in P. aeruginosa isolate from Japan, and then reported in Serratia marcescens isolate in Japan in 1991, IMP-2 was observed in Italy in A.baumannii. Currently there are 37 known IMP types which are most commonly seen in P. aeruginosa and A.baumannii isolates, but have also been reported in most Enterobacteriaceae members5. https://www.microrao.com/micronotes/pg/carbapenemases.pdf
One of the risk factors of acquiring carbapenem resistant bacteria is the increased consumption of carbapenems . Many patients are most likely affected by carbapenem resistant bacteria, including those who have poor immunity, prolonged hospitalization, admission to ICU, an indwelling medical device, and multiple exposures to different antibiotics. Also the increased frequency of international travel for work, leisure and migration contributes to their spread, and most of the carbapenemases are found on transferable plasmids, which are highly transmissible6. https://www.safetyandquality.gov.au/wp-content/uploads/2013/12/MRGN-Guide-Enterobacteriaceae-PDF-1.89MB.pdf
The prevalence of carbapenemase producing gram negative bacteria is increasingly reported in Sudan and Africa. Manenzhe et al. (2014) reported about 83 studies conducted in Africa which showed that the prevalence of carbapenemase producer isolates in hospital settings ranged from 2.3% to 67.7% in North Africa and from 9% to 60% in sub-Saharan Africa. Oxacillinases especially blaOXA48 was the most predominant in the whole country7.
In Sudan there are limited reports of carbapenemase producers, therefore the present study was performed8. This study aimed to detect carbapenem resistant Gram-negative bacilli (using conventional biochemical tests and chromogenic agar media), determine the antimicrobial susceptibility pattern of the isolates against carbapenems, detect carbapenemase enzyme production for the resistant isolates (using phenotypic methods, Modified Hodge Test), and detect blaOXA 48 and blaIMP genes for carbapenem resistant isolates (using PCR, genotypic methods).
This was cross sectional laboratory based study, conducted in Khartoum state in the period from February to August 2016. Ethical clearance was obtained from SUMASRI (University of Medical Sciences and Technology Sudanese Institute of Medical and Scientific Research) Institutional Review Board (SIRB); (IRB No: 00008867), which ensures that all ethical considerations for conducting the research in a way that protects patient’s confidentiality and privacy are followed. Informed consent was obtained from the hospital laboratories (laboratory manager) after providing them with the ethical clearance to collect samples during routine procedures from the microbiology laboratories. Participants’ privacy and confidentially was protected for all samples; personal information was not of great value in the current study and was thus not taken.
One hundred and forty nine Gram-negative bacilli were isolated from 147 different clinical specimens that were collected from patients attending different hospitals in Khartoum state. Blood agar, MacConkey agar, CLED media, and XLD media were used for primary plating depending on the type of clinical specimens, cultures were examined macroscopically for colonial morphology, and Gram stain was performed from suspected colonies. All Gram-negative bacilli isolates were selected then subcultured on MacConkey agar media for purity and further identification tests, and incubated at 37° overnight. Chromogenic agar media and different standard biochemical reactions, including oxidase, Kligler Iron Agar (KIA), urease production, citrate utilization, indole production, and motility tests were performed for their identification (all media mentioned above were obtained from LAB M, UK, except chromogenic media which was obtained from Laboratories Flow Media, Sudan)9.
Antimicrobial susceptibility testing to carbapenem antibiotic was performed for all Gram-negative bacilli isolates using the disc diffusion method, according to the Clinical Laboratory Standards Institute Guidelines9. Bacterial colonies were suspended in sterile normal saline and compared with McFarland standard and cultured in Muller-Hinton agar media (media were obtained from Pronadisa Laboratories Conda., Spain) using a sterile cotton swab. After overnight incubation at 37°C, zone of inhibition was measured and the reading was compared to the sheet provided by manufacturer9.
Gram-negative isolates showing resistant zones to Meropenem were tested for carbapenemase enzyme production using MHT and PCR.
MHT was used to detect carbapenemase enzyme production10. McFarland 0.5 dilution of E.coli (ATCC 25922) was prepared in 5 ml of sterile saline. The suspension was diluted to 1:10 by adding 0.5 ml of the 0.5 McFarland (Oxoid, UK) to 4.5 ml of sterile saline. A lawn of the 1:10 dilution of E.coli was streaked onto a Mueller Hinton agar plate using a sterile cotton swab. A 10 μg Meropenem susceptibility disk (Hi media, India) was placed on the center of the test area. The tested bacilli was streaked in a straight line from the edge of the disk to the edge of the plate. The plate was incubated overnight at 37°C in ambient air (Figure 1)11.
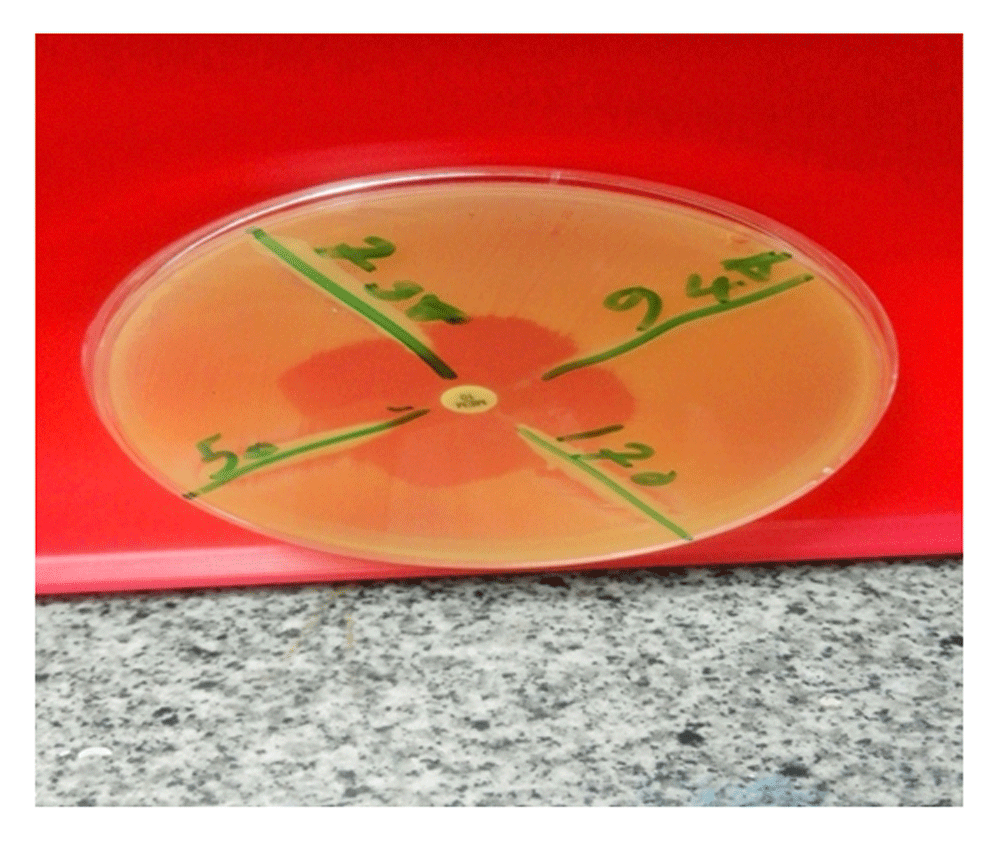
Clover leaf like indentation from E. coli ATCC showing positive result.
Positive MHT: Shown by clover leaf-like indentation of E. coli susceptible strains growing along the tested organism growth streak within the disk diffusion zone (Figure 1)12.
Negative MHT: No growth of E. coli along the tested organism growth streak within the disc diffusion zone12.
DNA extraction. DNA extraction for all resistant isolates was done by boiling method from fresh bacterial cultures within 24 hours, in which bacteria are in the logarithmic growth phase, which is the most suitable phase for bacterial DNA extraction according to the following protocol: Cells of interest were suspended in sterile normal saline and pelleted (106 to 107) by centrifugation at 4.000 rpm for 5 minutes using a labelled 1.5 ml safe-lock tube. The pellet was resuspended in 100 μl of phosphate buffer saline. The tubes were placed at 95°C for 15 minutes then centrifuged at >10.000 rpm for 5 minutes to pellet the cellular debris. Then the supernatant (lysate) was transferred into a labelled 1.5 ml safe-lock. The lysate was stored at -20 to 4°C. https://www.dkfz.de/gpcf/fileadmin/ccontrol/lysate_Protocol_DKFZ.pdf
PCR. A multiplex conventional PCR was designed to detect blaIMP and blaOXA 48 using specific primers for each. The primers were obtained from Macrogen Company, Korea (Table 1).
| Primer | Sequence 5′ - 3′ | Length (bp) |
|---|---|---|
| BlaOXA-48-F BlaOXA-48-R | GCTTGATCGCCCTCGATT GATTTGCTCCGTGGCCGAAA | 238 |
| BlaIMP–F BlaIMP-R | TCGTTTGAAGAAGTTAACGG ATGTAAGTTTCAAGAGTGATCC | 568 |
The amplification of DNA was performed using a TC-312 PCR machine (TECHNE, UK). For amplification, 17 μl of distilled water were added to ready manufactured premix solution (Intron biotechnology, Korea) and mixed well. Then 0.5 μl from forward primer and 0.5 μl from reverse primer of both blaIMP and blaOXA48 genes were added. Then 2 μl of DNA was added to the mixture. For both genes, the cycling conditions were: initial denaturation at 94°C for 5 minutes, denaturation at 94°C for 30 seconds, annealing at 53°C for 1 minute, and elongation at 72°C for 1 minute. The cycles were repeated 35 times, with a final extension step at 72°C for 5 minutes13. PCR product (5μl) were analyzed by gel electrophoresis in 1.0% agarose stained with ethidium bromide. The results were photographed under ultraviolet light machine (Transillumnator; Uvite, UK) to detect the specific amplified product by comparing it with 100 base pairs standard DNA ladder (Figure 2)14.
One hundred forty seven different clinical specimens were collected from patients attending different hospitals in Khartoum state. One hundred forty nine Gram-negative bacilli were isolated. Urine specimens were the most frequent specimen, comprising 104 out of 147 clinical specimens; 106 Gram negative bacilli were isolated from the 104 urine specimens.
The isolated Gram-negative bacilli comprised of 81(54.4%) E.coli, 44 (29.5%) Klebsiella species, 17(11.4%) Proteus species, 6(4.0%) Pseudomonas species and 1(0.7%) Enterobacter species (Table 2).
carbapenem (Meropenem) susceptibility testing using disc diffusion method showed that 75 (50.3%) of Gram-negative rods isolates were Meropenem resistant, 57 (38.3%) were Meropenem sensitive, and 17 (11.4%) were Meropenem intermediate (Table 3).
Regarding carbapenem (Meropenem) resistance, the most resistant organism was E. coli, which constituted 45 out of 75 resistant bacteria (30.2%), followed by Klebsiella species 15.4% (23 isolates), Proteus species 2.7% (4 isolates), Pseudomonas species 1.3% (2 isolates), and Enterobacter species 0.7% (1 isolate) (Table 3).
carbapenemase production by both MHT and PCR constituted 42 isolates (56%) of the total carbapenem resistant isolates.
MHT was performed for the 75 resistant isolates, 42 isolates (56%) were positive and the other 33 (44%) were negative for carbapenemase enzyme production by MHT (Table 4).
Conventional PCR assay for detection of blaOXA 48 and blaIMP genes was performed for all 75 resistant isolates. 17.3% of the resistant Gram-negative isolates were positive for blaOXA 48 gene, while 6.7% of them were positive for blaIMP gene; both constituted 24% carbapenemase producing bacteria using PCR (Table 5).
carbapenemase production constituted 42 isolates (56%) of all resistant isolates by both MHT and PCR; some were positive by both methods, some were positive by MHT only, and others were positive by PCR only.
Regarding MHT, E.coli was the most carbapenemase producer among the resistant isolates (it constituted 36% of the 56% resistant isolates) followed by Klebsiella species (it constituted 16% of the 56% resistant isolates). Pseudomonas species and Enterobacter species constituted 2.7% and 1.3%, respectively (Table 4).
Regarding blaOXA48 gene production, E.coli was the organism that harboured the blaOXA48 gene the most, which constituted 12.0% of the 17.3% blaOXA48 gene positive result, followed by Klebsiella species (4%), and Pseudomonas species (1.3%). Proteus species and Enterobacter species were negative for blaOXA48 gene. blaIMP gene was positive only for E.coli (constituted the whole percentage of resistant bacteria that harbored blaIMP gene). None of the other types of resistant Gram-negative isolates harboured the blaIMP gene (Table 5).
In this study E.coli was the most predominant organism among the isolated Gram-negative bacteria followed by Klebsiella species. This result agreed with the study done by Hayajneh, et al. in Jordan 2011–2013, and disagreed with the study done by Mataseje, et al. in Canada 2009–2010, in which Pseudomonas species was the commonest isolate14,15. This may be due to the difference in sample size and study area.
The resistance to carbapenem antibiotics in this study was 50.3%, which is not in agreement with a previous study conducted in India by Henkhoneng Mate, et al., the study conducted by Hayajneh, et al. in Jordan, and the study conducted by Gladstone et al., which showed that resistance to carbapenem antibiotics was 30%, 1.6%, and 12.2% respectively2,13,16. This may be due to the uncontrolled and misuse of antibiotics including broad spectrum antibiotics in our study area (Sudan)17.
Urine samples had the maximum number of carbapenem resistant isolates, which was similar to the study conducted by Henkhoneng Mate, et al in India2.
carbapenemase production constituted 42 isolates (56%) of the total carbapenem resistant isolates, which disagreed with the study conducted in Tanzania 2013. That study showed that 35% of the resistant isolates were carbapenemase producers; this may be to the difference in the methods used for enzyme detection, since the previous study used only genotypic methods, while the present study used both phenotypic and genotypic methods14. A study conducted in India 2015 by Panduragan, et al reported that 62% of isolates produced carbapenemases, found using both phenotypic and genotypic methods, which is agreed with the current study, which reported a near percentage (56%)12.
In this study, the most carbapenemase producing organism was E. coli, which disagrees with a previous study done by Mushi et al, who reported that K. pneumoniae was the most predominant carbapenemase producer14.
Regarding MHT, 50.6% of the total carbapenem resistant isolates were positive for carbapenemase production. This was not in agreement with a study conducted in India 2015 by Pandarangan, et al, who reported lower percentage (30.5%), and also disagreed with the study conducted by Henkhoneng Mate, et al in the same country, which reported a higher percentage (60.4%)2,12. This may be due to the difference in study area, which may affect the types and percentage of carbapenemase enzymes according to their spread.
Regarding blaOXA48 gene, 17.3% of the isolates harboured this gene, which disagreed with a previous study done by Memish, et al in Saudi Arabia 2015. This previous study showed a higher percentage (61.3%) of isolates with the blaOXA48 gene, which may be due to the different study area18.
blaIMP gene was harboured by 6.7% of the resistant isolates, which also disagreed with the study done in Tanzania (2013) and Thailand (2008). These studies showed that 21.6% and 15.4% of the resistant isolates, respectively, harboured the blaIMP gene14,19. In addition, a study performed by Nasr El-din in Egypt 2014 revealed that the blaIMP gene was absent from isolates20. This may be due to the different study area, the difference in sample size and the five different PCR assays that were performed in the study done in Tanzania, thus increasing the sensitivity and specificity compared to the present study.
In this study, blaOXA 48 gene was the most predominant carbapenemase harboured by the resistant isolates. This is in agreement with the study performed by Memish et al in Saudi Arabia 201518. However, our results disagreed with the study performed in Tanzania 2013, which showed that blaIMP gene was the most predominant14. This may be due to the difference in sample size and the five different PCR assays that were performed in this study.
This study concludes that the percentage of resistance to carbapenems in Khartoum state, Sudan, is very high and must be taken in consideration by the Sudanese government, and any large organization that can help in the control and prevention of carbapenemases, such as the World Health Organisation. E. coli, followed by Klebsiella species, were the most carbapenem resistant organisms and the largest carbapenemase producers. MHT is a simple method that can detect many types of carbapenemase enzymes, but it cannot detect some types of and does not specify the type of enzymes produced. In contrast, PCR is more sensitive, rapid and specific method for detection of specific types of carbapenemase enzymes. Additionally in the present study, the percentage of Gram-negative bacilli that produce blaOXA48 gene was more than those producing blaIMP; however, one of the isolates harboured both blaOXA48 and blaIMP genes. In this study, some Gram-negative bacilli were positive for carbapenemase enzyme production by both MHT and PCR, some were negative by MHT and positive by PCR, and some of them were positive by MHT and negative by PCR (this study did not detect all carbapenemase genes using PCR; thus those positive by MHT but negative by PCR may process other type of carbapenemase enzymes rather than OXA48 and IMP. Although this may also be because the gene is not expressed).
A larger sample size should be tested to cover a wider range of isolates.
Other specific tests for detection of carbapenemase enzymes should be used, such as EDTA disc synergy test, MDI, RDS, MBL E test, more primers should be used to detect most types of carbapenemase enzymes using PCR, and different types of PCR assays should be coupled with each other to increase the sensitivity for enzyme detection.
Detection of carbapenemase producers should be introduced as routine tests in microbiology labs for rapid detection of resistant isolates and to control their spread, especially for newly admitted patients to the hospitals.
Prevention and control programs of carbapenem resistant Gram-negative bacteria should be performed to prevent the spread of carbapenemase producers, which includes appropriate use of antimicrobials and facility-level prevention strategies, as recommended by the CDC.
Dataset 1: Raw data for the 147 specimens analysed showing the results of the genotypic and phenotypic tests. The file contains the same data in Excel and SPSS formats. DOI, 10.5256/f1000research.12432.d17637721.
Dataset 2: Pictures related to the methods used in the current study (in zipped file): ‘Standard antimicrobial susceptibility testing’, ‘Modified Hodge Test (MHT)’ and ‘Molecular detection of blaIMP and blaOXA 48 genes’. DOI, 10.5256/f1000research.12432.d17637822.
I would like to offer my special thanks to Miss Maha Baballah for her assistance especially in sample collection. Also Dr. Abd Elhakam Hassan Ibrahim for his strong assistance in keeping my progress in the practical work (especially MHT). My grateful thanks are also extended to Dr. Asim Halfawi for his contribution to the analysis of the data.
My great appreciation and thanks are extended to those who helped me during my practical work; Eman Eshag, Tahani Mursal, Roaa Malik, Samah Abdallah, Reham Abdelrahman, and Mogadam Bahr Eldeen who helped me a lot in susceptibility testing and DNA extraction. Khansa Esam who helped me in sample collection and culture technique, Hyam Abdelrahman and Aymen Jalal Eldeen who assisted in the software. I would like to thank the lab staff of Sudan University of Sciences and Technology for giving me the chance to perform the PCR assay in their laboratory and assisting me.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
I cannot comment. A qualified statistician is required.
Are all the source data underlying the results available to ensure full reproducibility?
Partly
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Microbiology
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
No
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Partly
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Bacterial resistance in Gram negative.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 07 Sep 17 |
read | read |
Click here to access the data.
Spreadsheet data files may not format correctly if your computer is using different default delimiters (symbols used to separate values into separate cells) - a spreadsheet created in one region is sometimes misinterpreted by computers in other regions. You can change the regional settings on your computer so that the spreadsheet can be interpreted correctly.
Click here to access the data.
Spreadsheet data files may not format correctly if your computer is using different default delimiters (symbols used to separate values into separate cells) - a spreadsheet created in one region is sometimes misinterpreted by computers in other regions. You can change the regional settings on your computer so that the spreadsheet can be interpreted correctly.
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)