Keywords
multiqc, conformation, saccharides, visualisation, macrocycles
multiqc, conformation, saccharides, visualisation, macrocycles
Cyclic molecules and systems consisting of cyclic molecules play central roles in several chemical and biological processes1,2. RNA, DNA, proteins and carbohydrates are polymers that all contain cyclic moieties that exhibit wide ranging conformational variety. For example, the puckering of fructose can be used to illustrate the value of determining conformational statistics as recognition and binding to Fructokinase is due to pucker specificity. Similarly different sugar puckers of ribose in RNA and de-oxyribose in A-DNA and Z-DNA account for the molecular shape of the respective nucleotide helices2. In DNA-protein complexes any kink in the DNA axis is accompanied by a change in deoxy-ribose pucker, demonstrating the important connection pucker plays in molecular structure and flexibility3,4. In enzymatic transition states for glycosyl hydrolases and transferases, conformations change to provide the correct electronic state for the reaction to take place5.
Conformational analyses, which recognise shapes (Figure 1), are important for finding transition state leads, understanding molecular recognition events, ligand fitting in macrocycles and for molecular pattern matching.
We created a standardized toolset called Tessellate & Montage for comparisons of cyclic molecule conformations (ring pucker) from structural databases and simulation trajectory data. Tessellate can readily find, calculate and quantify cyclic conformations of cycles and macrocycles in datasets. Montage forked from MultiQC6 is a transferable reporting package for visualizing the conformations of cycles. Montage addresses the need to compare multiple molecular data sets by incorporating results from multiple analyses into a single report. Similar tools for calculating ring conformations are shown in Table 1, however these tools do not provide methods for calculating the ring pucker of macrocycles, nor do they have the reporting capability for comparing multiple types of molecular data sets in a single report. Existing pucker models include perpendicular displacements from a mean plane7,8, intracyclic torsions9,10 and triangular tessellation11–15.
| Resource | Open source/Free | Webapp | Details |
|---|---|---|---|
| RING16,17 | Yes, requires password. | No | CP pucker. |
| Cremer-Pople (CP) parameter calculator | Yes | Yes | CP pucker. |
| VMD18, paper chain representation19 | Yes | No | In VMD. CP pucker. |
| GlyTorsion | Yes | Yes | PDB statistics using ring torsion angles. |
| CHARMM CORREL command20 | No | No | In CHARMM. CP Pucker timeseries. |
A further aim was to ensure that these tools (Tessellate and Montage) function independently from informatics workflows systems, while also being readily incorporated and fully functional as part of systems such as Galaxy21 and the Glycome Analytics Platform22.
Tessellate is a package for quantifying ring conformation and qualifying the conformer in terms of a canonical conformer (e.g. chair, boat). Developed in Python, Tessellate uses BioPython23 for accessing protein data bank files. Using Tessellate with Visual Molecular Dynamics (VMD) is possible through a tcl script provided. Visualising the results of this analysis is done using Montage.
Montage is a fork of the excellent MultiQC6 and provides reporting templates and functionality for computational chemistry and chemical glycobiology. Previously we prototyped reporting within a web platform, the aim here was to ensure that the visual reports connected easily into workflow systems such as Galaxy21, GAP22 and were also transferable outside of these platforms.
These tools were designed to work in Linux.
Triangular tessellation for ring sizes 5–8 – Implemented as per the following papers11,14,15. Protein and water residues in the input structures are ignored by default.
Nomenclature and ring ordering – Conformational qualification depends on a consistent ring ordering according to the IUPAC definition24. Residues atoms are matched to an internal ligand dictionary and aligned in best-possible cyclic order to prevent atom ordering mistakes.
Ring finding algorithm – Cycles are detected using the Smallest Set of Smallest Rings (SSSR) ring perception algorithm25.
Conformational quantitation – Coordinates are matched to known canonical conformers coordinates. The best match is returned.
Macrocyclic – Macrocycle conformations are returned based on the centre of geometry of any subsidiary cycles.
Requirements: Python 3, Linux. For PDB analysis, BioPDB is required. Detailed installation instructions, package requirements and usage examples are provided in the README file of the released software (see Software availability section).
Tessellate reads in molecular data and outputs a summary of cyclic conformations (Input file types: list containing PDB IDs, PDB file, list of atomic coordinates. Output file types: JSON, txt). Montage reads the summary from Tessellate and produces a html report with a summary chart and charts for 5,6,7,8 rings found in the inputs (Input file types: JSON, txt. Output file types: HTML).
Usage example:
The workflow is as follows:
1. Choose molecular data e.g. from the PDB or use VMD to create a list of atomic coordinates from molecular simulation data.
2. Run Tessellate on the Linux command line e.g. `tessellate data/usecase-*DNA --input-format=pdblist --output-format=json --output-dir=output-usecase-rnadna`.
3. Run Montage on the Linux command line e.g. `multiqc output-usecase-rnadna -m montage_tessellate`.
4. Open report using a web browser.
Simulation – Ribose in vacuo biased molecular dynamics calculation
The conformational changes during a biased free energy simulation for ribose. This analysis can be used to identify patterns in conformational change and confirm adequate sampling of phase space (Figure 2). VMD was used to open and read coordinate and trajectory files, and write out the atomic coordinates of the ribose 5 membered ring. The atomic coordinates are read in by Tessellate, producing pucker coordinates stored as JSON. MultiQC creates an HTML report from the JSON input.
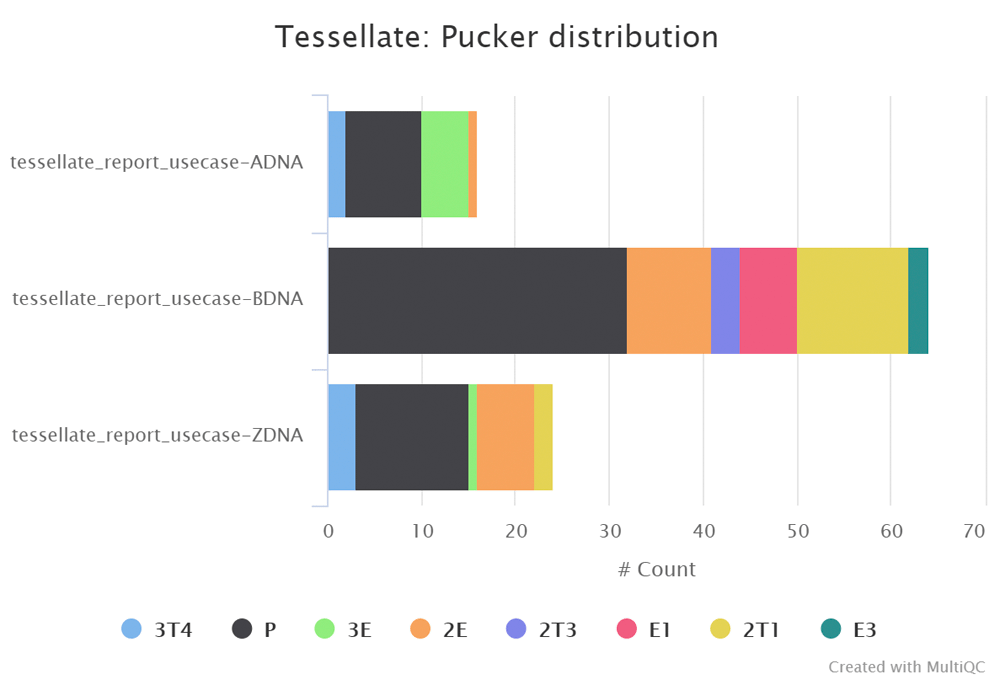
Comparative conformational character of DNA and RNA
The nucleotides A-DNA and Z-DNA tend to be C3’-endo (3E), but in B-DNA the tendency is to C2’-endo (2E); this accounts for the different molecular shape of the respective nucleotide helices. To explore this, PDB structures that are representative of A, B and Z DNA (1ANA, 3BSE, and 2DCG) were analysed (see Operation section and Figure 3).
Cyclodextrins
An analysis of alpha cyclodextrin (ACD) from the PDB shows the glucose monomer conformations are mostly 4C1 (85.9%) with few deviations (Figure 4). The ring pucker of the macrocycle, as defined by treating the centre of geometry for each glucose in cyclodextrin as an atom, varies between planar (P) and half chair (H, e.g. 3H4, 5H4,2H3) conformations.
Results from the three use cases can be found in Supplementary File 1.
Tessellate enables users to compile datasets from the reservoir of PDB and simulation produced data. From this ensemble of experimentally and computationally determined ring structures, the relation between cyclic conformational preference and other variables can be discovered using Montage.
Abbreviations: ACD: Alpha Cyclodextrin; GAP: Glycome Analytics Platform; JSON: JavaScript Object Notation; PDB: Protein Data Bank; SSSR: Smallest Set of Smallest Rings
Latest source code:
Tessellate: https://github.com/scientificomputing/Tessellate
Montage: https://github.com/scientificomputing/Montage
Archived source code as at the time of publication:
Tessellate: https://doi.org/10.5281/zenodo.106865626
Montage: https://doi.org/10.5281/zenodo.106869227
Tessellate License: Apache 2.0
Montage License: GNU GPLv3
This publication is based on research that has been supported in part by the University of Cape Town’s Research Committee and the National Research Foundation of South Africa (NRF; grant 87956). This work is based in part upon research supported by the South African Research Chairs Initiative of the Department of Science and Technology and the NRF (grant 48103).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Supplementary File 1: Zip file containing Tessellate and Montage outputs for the 3 use cases:
tessellate_report_usecase-timeseries.json
tessellate_report_usecase-ADNA.json
tessellate_report_usecase-BDNA.json
tessellate_report_usecase-ZDNA.json
tessellate_report_usecase-ACD.json
multiqc_report_usecase1.html
multiqc_report_usecase2.html
multiqc_report_usecase3.html
Click here to access the data.
The datasets are kept at https://github.com/scientificomputing/tessellate/tree/master/data (see Software availability section).
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the rationale for developing the new software tool clearly explained?
Yes
Is the description of the software tool technically sound?
Yes
Are sufficient details of the code, methods and analysis (if applicable) provided to allow replication of the software development and its use by others?
Yes
Is sufficient information provided to allow interpretation of the expected output datasets and any results generated using the tool?
Yes
Are the conclusions about the tool and its performance adequately supported by the findings presented in the article?
Yes
References
1. Foley BL, Tessier MB, Woods RJ: Carbohydrate force fields.Wiley Interdiscip Rev Comput Mol Sci. 2012; 2 (4): 652-697 PubMed Abstract | Publisher Full TextCompeting Interests: No competing interests were disclosed.
Is the rationale for developing the new software tool clearly explained?
Yes
Is the description of the software tool technically sound?
Yes
Are sufficient details of the code, methods and analysis (if applicable) provided to allow replication of the software development and its use by others?
Yes
Is sufficient information provided to allow interpretation of the expected output datasets and any results generated using the tool?
Yes
Are the conclusions about the tool and its performance adequately supported by the findings presented in the article?
Yes
Competing Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 08 Dec 17 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)