Keywords
Plasmodium falciparum, Toxoplasma gondii, drug targets, essential genes
Plasmodium falciparum, Toxoplasma gondii, drug targets, essential genes
Malaria killed an estimated half a million people in the year 2015, 70% of them were children under the age of five1. The emergence and spread of Plasmodium falciparum strains resistant to all currently used anti-malarial drugs2 has created an urgent need to discover new drugs. New anti-malarial drugs are also needed for malaria elimination and global eradication, for which the currently available drugs are not adequate3. There are two main approaches for drug-discovery against pathogens: Phenotype screening and target-based approach4. In phenotype screening, compounds are identified that inhibit the cellular growth of the pathogen. Large-scale screening of millions of compounds against the erythrocytic stage of P. falciparum has identified thousands of such inhibitors5. Some of these inhibitors have progressed to clinical trials6. In the target-based approach, compounds are identified that inhibit the activity of a protein essential for the viability of the pathogen. Thus target-based approach requires previous knowledge about genes that are essential for the pathogen. Only a few essential genes have been identified in P. falciparum, hampering the target-based approach for anti-malarial drug discovery. Consequently, target-based approach has only identified a few anti-malarial candidates6. However, recent large-scale screening of about 2500 genes in a rodent malaria parasite P. berghei has identified about 1200 essential genes7,8. A recent genome-scale CRISPR screen in a related apicomplexan parasite Toxoplasma gondii has identified about 3000 essential genes9. Here we analyse this data and find significant conservation of gene essentiality in these two pathogens. From this, we identified potential anti-malarial drug targets that exhibit conserved essentiality in apicomplexan parasites; we predict novel essential genes in Plasmodium based on the essentiality of their orthologs in T. gondii. These targets could serve as starting points for target-based anti-malarial drug discovery.
The genome-wide CRISPR screening data on the relative fitness of T. gondii genes during infection of human fibroblasts cells was obtained from Sidik et al.9. The authors defined log2 fold change in abundance of single guide RNA (sgRNA) targeting a given gene as the “phenotype” score for that gene9. It was found that for a previously determined set of 81 essential and non-essential genes, a phenotype score of less than -2 identified most of the essential genes, but none of the non-essential genes9. We thus defined all genes with a phenotype score of less than -2 as essential (2870 genes). Genes with a phenotype score greater than 0 were defined as non-essential (3071 genes), while those with a phenotype score between 0 and -2 were not classified (2210 genes). The in vivo relative growth rate data for 2574 genes of P. berghei were obtained from the PlasmoGEM database7,8 (http://plasmogem.sanger.ac.uk/phenotypes). The authors generated knockout mutants by transfection with large pools of barcoded gene knockout vectors. The in vivo growth rate in Balb/c mice was obtained by counting barcodes by next generation sequencing daily between days 4 and 8 post transfection7. Essential genes were defined as genes with a growth rate not significantly different from 0.1 (growth rate of the wild type taken as 1), while non-essential genes were defined as genes with growth rate not significantly different from 17.
Proteome sequences of P. falciparum 3D7, P. berghei ANKA, P. chabaudi chabaudi, P. cynomolgi B, P. knowlesi H, P. reichenowi CDC, P. vivax Sal1, P. yoelii 17X were downloaded from the PlasmoDB database10 (http://plasmodb.org/common/downloads/release-27/). The Proteome sequences for six apicomplexan species were obtained from EuPathDB11: Cryptosporidium hominis TU502 (http://cryptodb.org/common/downloads/release-29/ChominisTU502/); T. gondii GT1 (http://toxodb.org/common/downloads/release-29/TgondiiGT1/); Eimeria brunetti Houghton (http://toxodb.org/common/downloads/release-29/EbrunettiHoughton/); Babesia bovis T2Bo (http://piroplasmadb.org/common/downloads/release-29/BbovisT2Bo/); Theileria annulata Ankara (http://piroplasmadb.org/common/downloads/release-29/TannulataAnkara/); and Gregarina niphandrodes (http://cryptodb.org/common/downloads/release-29/GniphandrodesUnknown/). Proteome sequences for Homo sapiens were downloaded from EBI (http://www.ebi.ac.uk/reference_proteomes). Homologs of P. berghei genes in H. sapiens were identified with E-value cut-off of 1e-6, with soft mask set as true. Orthologous sequences were identified using best bidirectional hit algorithm12.
RNA-seq data (FPKM values) for different stages of P. berghei was obtained from Otto et al.13. Proteomics data on different stages of P. berghei and dN, dN/S values were obtained from Hall et al.14. Gene Ontology information for P. falciparum was obtained from PlasmoDB10, and these functions were assigned to their orthologous proteins in P. berghei. Enzyme Commission (EC) numbers for P. berghei and P. falciparum were also obtained from PlasmoDB. Trans-membrane regions were identified using TMHMM15. All statistical analyses were performed in the R software version 3.3.1 (https://www.r-project.org/).
The relative in vivo growth rate of knockout mutants for 2574 P. berghei genes (out of total 5076 genes in P. berghei) has recently been measured, of which 1198 genes (46%) with very low growth rate were classified as essential7,8. Similarly, in vivo relative fitness of knockout mutants for 8151 T. gondii genes have been measured9, of which 2870 genes (35%) with very low relative fitness values were classified as essential (see Methods). Of the 2574 P. berghei genes with fitness data, 1617 genes have an ortholog in T. gondii. P. berghei genes with an ortholog in T. gondii were significantly more likely to be essential, compared to P. berghei genes without an ortholog in T. gondii (53% vs. 36%; Fisher test p = 7e-18; Figure 1A). P. berghei genes with an essential ortholog in T. gondii were significantly more likely to be essential, compared to P. berghei genes with a non-essential ortholog in T. gondii (71% vs. 17%; Fisher test p = 6e-59; Figure 1A). There was a significant correlation in relative fitness values of P. berghei and T. gondii (Spearman correlation coefficient 0.47; p = 3e-89; n =1617; Figure 1B). The essentiality of 2502 P. berghei genes was not tested, but the essentiality information of T. gondii orthologs may be used to predict their essentiality in P. berghei. There were 687 genes in P. berghei with an essential ortholog in T. gondii, and thus may be predicted as essential in P. berghei (Dataset 116).
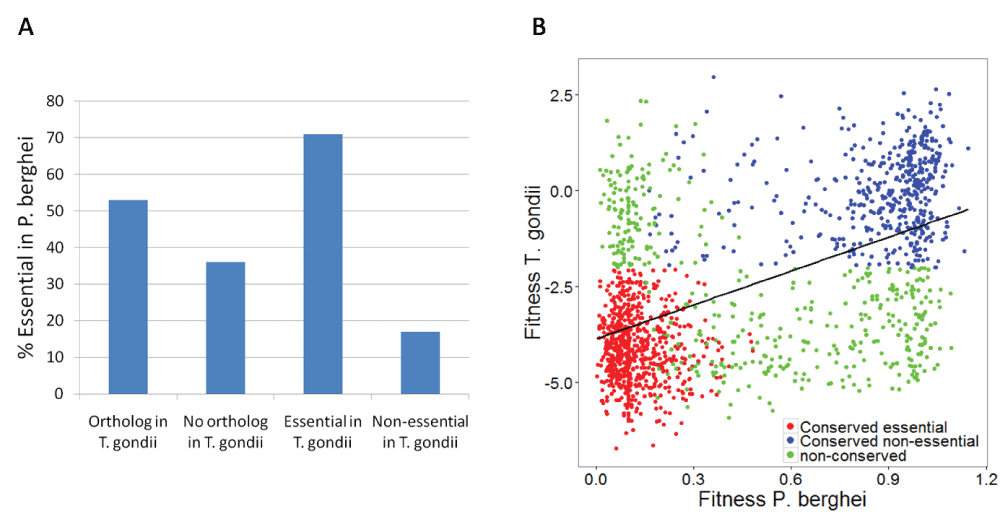
(A) P. berghei genes with an ortholog in T. gondii were more likely to be essential, compared to P. berghei genes without an ortholog in T. gondii (Fisher test p = 7e-18). P. berghei genes with an essential ortholog in T. gondii were significantly more likely to be essential compared to P. berghei genes with a non-essential ortholog in T. gondii (Fisher test p = 6e-59). (B) There was a significant correlation in relative fitness values of P. berghei and T. gondii (Spearman correlation coefficient 0.47; p = 3e-89; n =1617). Genes classified as essential in both species are colored red. Genes classified as non-essential in both species are colored blue. Genes that are essential in only one of the species are colored green.
We argue that genes identified as essential in both the apicomplexan parasites could be more useful drug targets for the following reasons: 1) Genome-scale fitness screens often involve significant false positives and false negatives7, thus genes identified as essential in independent experiments in different parasites could be more confidently assigned as essential; 2) the substantial conservation of gene essentiality between the two parasites demonstrates that essentiality information in T. gondii offers relevant information about gene essentiality in P. berghei; 3) genes that are essential in both P. berghei and T. gondii should be more likely to be essential in human malarial species, such as P. falciparum and P. vivax; 4) genes that are essential in both P. berghei and T. gondii should be more likely to be essential across different developmental stages of Plasmodium, which is a highly desirable property of Plasmodium drug targets17. We thus identified 710 genes that were essential in both species. A total of 289 of these 710 genes encode enzymes, which are typically used as drug targets against pathogens. Of these 289 genes, 245 had an ortholog in all Plasmodium species and did not have more than one trans-membrane segment. We removed proteins with more than one trans-membrane segments, as these are often difficult to purify for in vitro assays. Of the 245 proteins, 30 showed no significant sequence similarity to any human proteins (listed in Table 1), and 83 showed less than 30% identity and 151 showed less than 40% identity to any human protein (Dataset 116). Figure 2 shows the flow chart of the selection process.
Among the P. berghei enzymes that were not tested for essentiality, 186 had an essential ortholog in T. gondii and thus may be predicted as essential in P. berghei. To increase the confidence of these genes to be essential in Plasmodium, we considered 53 genes that were conserved across Plasmodium and apicomplexan species. Among the enzymes tested for essentiality, such a criteria led to a set with 77% enzymes as essential, suggesting high enrichment for essentiality among predicted essential enzymes. In total, 28 of these enzymes had low sequence similarity (<40% identity) with human proteins and thus may also be considered as potential drug targets (Dataset 116).
Essential genes show different expression, evolutionary and functional properties9. We thus tested whether similar patterns would be observed for P. berghei. Essential P. berghei genes showed higher mRNA expression levels in asexual stages, but lower expression levels in sexual stages compared to non-essential genes (Figure 3A). Proteins encoded by essential genes were more likely to be detected by mass-spectrometry in different developmental stages compared to non-essential genes (Figure 3B). Essential genes showed a lower evolutionary rate (dN and dN/dS) and higher conservation in apicomplexan species (Figure 3C). Essential genes were significantly enriched in functional classes, such as “Translation”, “Ribosome”, “DNA replication”, “Intracellular protein transport”, “Cytoplasm”, and “Nucleus” (Figure 4).
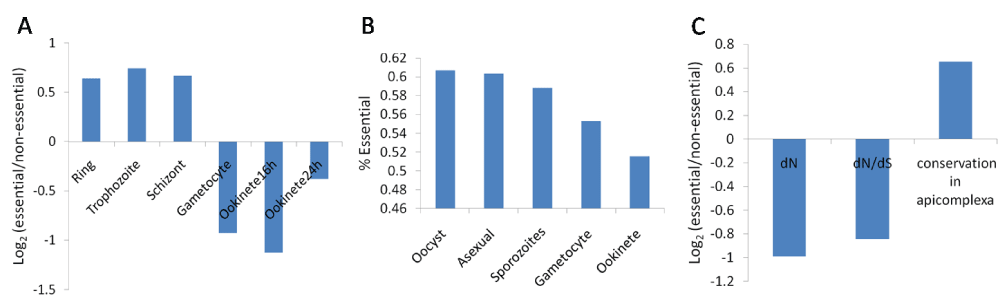
(A) Essential P. berghei genes showed higher mRNA expression levels in asexual stages, but lower mRNA expression levels in sexual stages. The mean FPKM values for the essential and non-essential genes were calculated for different development stages and their log2 ratio was taken. All stages except ‘ookinete 24h’ showed a statistically significant difference between essential and non-essential genes (t-test; p < 0.05). The RNA-seq data was taken from Otto et al.13. (B) Proteins encoded by essential genes were more likely to be detected by mass-spectrometry in different stages compared to non-essential genes. All stages except ‘sporozoites’ showed a significant difference between essential and non-essential genes (Chi-square test; p < 0.05). Overall 47% of the tested genes were essential. The proteomics data was obtained from Hall et al.14 (C) Essential genes showed a lower evolutionary rate and higher conservation across apicomplexan species. The mean dN and dN/dS values for essential and non-essential genes was calculated and their log2 ratio was taken. This data was taken from Hall et al.14. The mean number of apicomplexan species (out of six), in which an ortholog was identified, was calculated for essential and non-essential genes and their log2 ratio was taken. dN and conservation in apicomplexan species showed a statistically significant difference between essential and non-essential genes (t-test; p < 0.05), but not dN/dS.
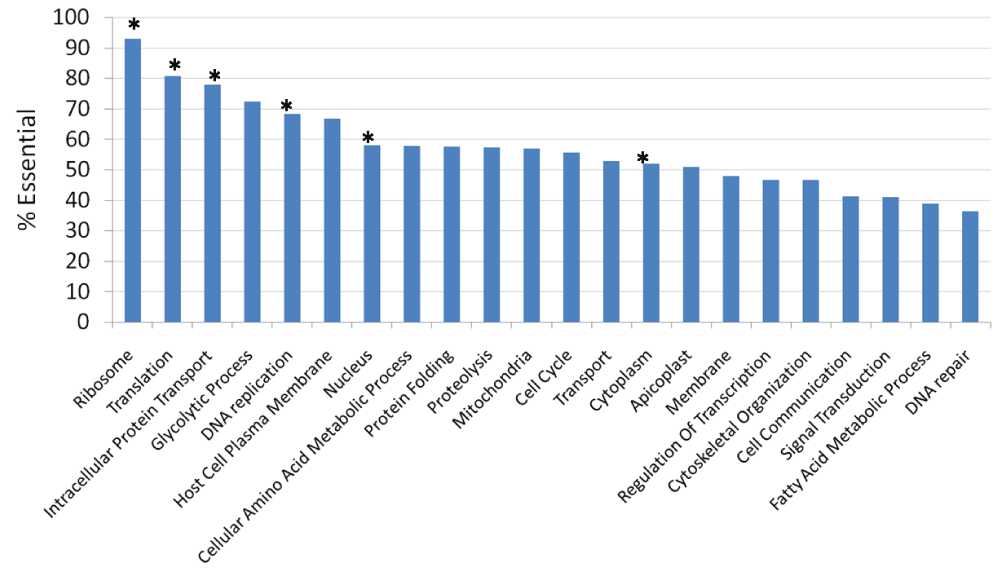
The Gene Ontology information for Plasmodium falciparum genes was obtained from PlasmoDB10 and assigned to their P. berghei orthologs. Classes with a significant difference (Chi-square test; p < 0.05) in essential genes are marked with *.
The recent availability of gene essentiality data from P. berghei and the related apicomplexan T. gondii provides an unprecedented opportunity to identify potential drug targets to accelerate anti-malarial drug discovery. We find a significant correlation of gene essentiality between P. berghei and T. gondii (Figure 1). Thus, the information about gene essentiality in T. gondii provides independent experimental support for gene essentiality in P. berghei, which not only increases the confidence of gene essentiality in P. berghei, but also increases the likelihood that these genes would be essential in other Plasmodium species that cause human malaria, and probably in different Plasmodium developmental stages. Drug targets that are essential in multiple species and stages of Plasmodium are particularly desirable17. Novel essential genes in Plasmodium could also be predicted based on the essentiality of their orthologs in T. gondii. Further prioritization of these genes could be made based on their conservation across Plasmodium and apicomplexan species, low sequence similarity to human proteins, as well as practical information, such as previous availability of clones, assays, protein structure and inhibitors18,19. The high conservation of essentiality between P. berghei and T. gondii may allow prediction of essential genes in other apicomplexan pathogens, such as Cryptosporidium.
We found gene and protein properties significantly associated with essentiality in P. berghei. At the mRNA level, essential genes, compared to non-essential genes, were expressed at higher levels in asexual stages, but at lower levels in sexual stages (Figure 3A). Since gene essentiality was measured at the asexual stage, this might explain the positive correlation between essentiality and mRNA expression in asexual stages. Proteins encoded by essential genes were more likely to be detected by mass-spectrometry in different development stages (Figure 3B). Essential genes showed lower evolutionary rates and higher conservation across apicomplexan species (Figure 3C). The higher evolutionary conservation of essential genes is well-documented20. We find Gene Ontology classes “Translation”, “Ribosome”, “DNA replication”, “Intracellular protein transport”, “Cytoplasm”, and “Nucleus” to be significantly enriched in essential genes (Figure 4). “Translation” class was also enriched in essential genes after excluding “Ribosome” genes (69% essential; Chi-square test; p = 0.0001), suggesting that enrichment of essential genes in the “Translation” category is not only due to ribosomal genes. Thus enzymes involved in protein translation may be important targets for anti-malarial drug discovery.
The in vivo relative growth rate data for 2574 genes of P. berghei genes was obtained from PlasmoGEM database (http://plasmogem.sanger.ac.uk/phenotypes)8. The genome-wide CRISPR screening data for the relative fitness of 8151 T. gondii genes during infection of human fibroblasts cells was obtained from Sidik et al.9.
Dataset 1: Fitness, expression, functionality, conservation and evolutionary information of Plasmodium berghei genes. doi, 10.5256/f1000research.10559.d14869816
G.P.S. conceived and designed the study, performed the research and wrote the manuscript.
The work is supported by an Early Career Fellowship to G.P.S. by the Wellcome Trust/DBT India Alliance (IA/E/15/1/502297).
The author would like to acknowledge suggestions and criticism on the manuscript by Ms. Preeti Goel.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
References
1. Crowther GJ: Referee Report For: Conservation of gene essentiality in Apicomplexa and its application for prioritization of anti-malarial drug targets [version 1; referees: 1 approved with reservations]. F1000Research. 2017; 6 (23).Competing Interests: No competing interests were disclosed.
References
1. Gomes AR, Bushell E, Schwach F, Girling G, et al.: A genome-scale vector resource enables high-throughput reverse genetic screening in a malaria parasite.Cell Host Microbe. 2015; 17 (3): 404-13 PubMed Abstract | Publisher Full TextCompeting Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 09 Jan 17 |
read | read |
Click here to access the data.
Spreadsheet data files may not format correctly if your computer is using different default delimiters (symbols used to separate values into separate cells) - a spreadsheet created in one region is sometimes misinterpreted by computers in other regions. You can change the regional settings on your computer so that the spreadsheet can be interpreted correctly.
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)