Keywords
Crustaceans, Neuropeptides, Crustacean Hyperglycemic Hormone (CHH), Transcriptomics, Comparative genomics
Crustaceans, Neuropeptides, Crustacean Hyperglycemic Hormone (CHH), Transcriptomics, Comparative genomics
Crustaceans and insects from the phylum Arthropoda have longstanding histories in peptide biology research, principally in areas related to the roles of peptide hormones in physiology and neuroendocrine signaling. Early discoveries have demonstrated that compounds in the crustacean nervous system were responsible for chromatophore control1–3. Four decades later, it was revealed that a compound known as the red pigment concentrating hormone functions as the first crustacean/invertebrate neuropeptide4. Since then, multiple studies have shed light on the highly pleiotropic functions of crustacean neuropeptides implicated in the regulation of a myriad of physiological processes such as light adaptation, molt inhibition, carbohydrate metabolism, reproduction and ion transport5–9.
The crustacean hyperglycemic hormone (CHH) represents a neuropeptide superfamily that is unique to arthropods6,10–12. This superfamily is made up of peptides containing ∼70 amino acids originally isolated from the X-organ-sinus-gland system of the decapod Carcinus maenas13. Given their high degree of structural similarities, and the conservation of six cysteine residues, the molt-inhibiting hormone (MIH) and gonad-inhibiting hormone (GIH) were considered as part of this family collectively known as CHH/MIH/GIH. To date, at least 150 CHH peptides have been isolated and characterized, mainly in decapods through comparative studies on endocrinology5–7,14–21. Although there are reports on CHH peptides in other crustacean taxa such as Armadillidium vulgare (Isopoda)22,23, Daphnia pulex (Cladocera)24 and Daphnia magna15, investigations beyond decapods have remained scant and the sequences of CHH/MIH/GIH genes in other crustacean taxa have remained elusive.
Here, we took advantage of the growing number of high-throughput crustacean datasets on public repositories to perform transcriptome mining of the CHH/MIH/GIH superfamily. To this end, we looked at crustacean species from three Classes (Figure 1) and annotated CHH/MIH/GIH genes. This high confidence set of genes identified using our in silico framework provides an important basis for understanding neuropeptide biology underpinning physiological adaptations across diverse crustacean species.
We retrieved complete transcriptome datasets for 112 crustacean species available at the time of manuscript preparation from the European Nucleotide Archive. Five non-crustacean arthropod proteomes were retrieved from Uniprot. A complete list of accessions used in this study is provided in Supplementary Table 1. We retrieved a list of query sequences used in subsequent homology searches from Uniprot and GenBank.
To identify CHH/MIH/GIH gene orthologs, we used multiple Basic Local Alignment Search Tool (BLAST)-based approaches such as BLASTp and tBLASTn with varying Blocks Substitution matrices based on a previously published workflow25. The BLAST results were filtered by e-value of < 10-6, best reciprocal BLAST hits against the GenBank non-redundant (nr) database and redundant contigs having at least 95% identity were collapsed using CD-HIT. We then utilized HMMER (version 3.1) employing hidden Markov models (HMM) profiles26 to scan for the presence of CHH Pfam domains27 on the best reciprocal nr BLAST hits to compile a final non-redundant set of crustacean CHH/MIH/GIH orthologs. Pfam annotations, associated e-values and fasta sequences are provided in Dataset 128 and Dataset 229.
Multiple sequence alignments of CHH protein sequences were performed using MAFFT (version 7)30. Phylogenetic tree was built from the MAFFT alignment using RAxML WAG + G model to generate best-scoring maximum likelihood trees31. Geneious (version 7) was used to generate multiple sequence alignment images as well as graphical representations of the Newick tree32.
We have annotated CHH/MIH/GIH genes from 112 crustacean transcriptome datasets representing three Classes: Malacostraca (Amphipoda: 56 species, Decapoda: 14 species, Isopoda: 27 species, Euphausiacea: 2 species and Mysida: 1 species), Branchiopoda (3 species), and Copepoda (9 species) (Supplementary Table 1). We also looked at 5 non-crustacean species from Arthropoda: Insecta (3 species), Arachnida (1 species) and Chilopoda (1 species) (Supplementary Table 1). Using sequence and motif similarity based approaches, we have conservatively identified a total of 413 genes from these transcriptomes (Figure 2; Dataset 128 and Dataset 229).

Heat maps denote the number of CHH/MIH/GIH genes identified from each crustacean species. CHH/MIH/GIH genes from five non-crustacean species within Arthropoda are also shown.
Multiple sequence alignment analyses on representative CHH/MIH/GIH sequences revealed the presence of a conserved set of six cysteine residues (Figure 3), likely contributing to the formation of disulfide bonds33. Comparison of insect sequences from Drosophila melanogaster, Anopheles gambiae and Aedes aegypti demonstrated sequence identities of at least 46% (Supplementary Table 2). Within crustacean taxa, a range of sequence identities were observed: Branchiopoda (∼25% to 93%), Copepoda (∼12% to 30%) and Malacostraca (∼10% to 98%) (Supplementary Table 2). This is reflected in the phylogeny where CHH/MIH/GIH sequences from related individuals form distinct clusters (Figure 4). It was previously reported that multiple gene duplications of CHH family peptides occurred in the decapod lineage leading to a high degree of genetic polymorphism15, hence providing an explanation for our current observation. Two separate clusters of CHH genes exhibiting antagonistic patterns of expression were identified in the decapod Metapenaeus ensis, posited to represent an ancient gene duplication event34. Although it is not possible to pinpoint the genomic loci of CHH sequences identified from this study, it is likely that paralogous copies offer mechanisms for evolving new functions through functional divergence. CHH-like genes arising from duplication of the ancestral copy are subjected to reduced selective pressure and therefore may lose their hyperglycemic activity to adopt more specialized roles15. Further biochemical studies will be required to unravel the functions of the novel genes identified from this study.

Six conserved cysteine residues are annotated within red boxes.
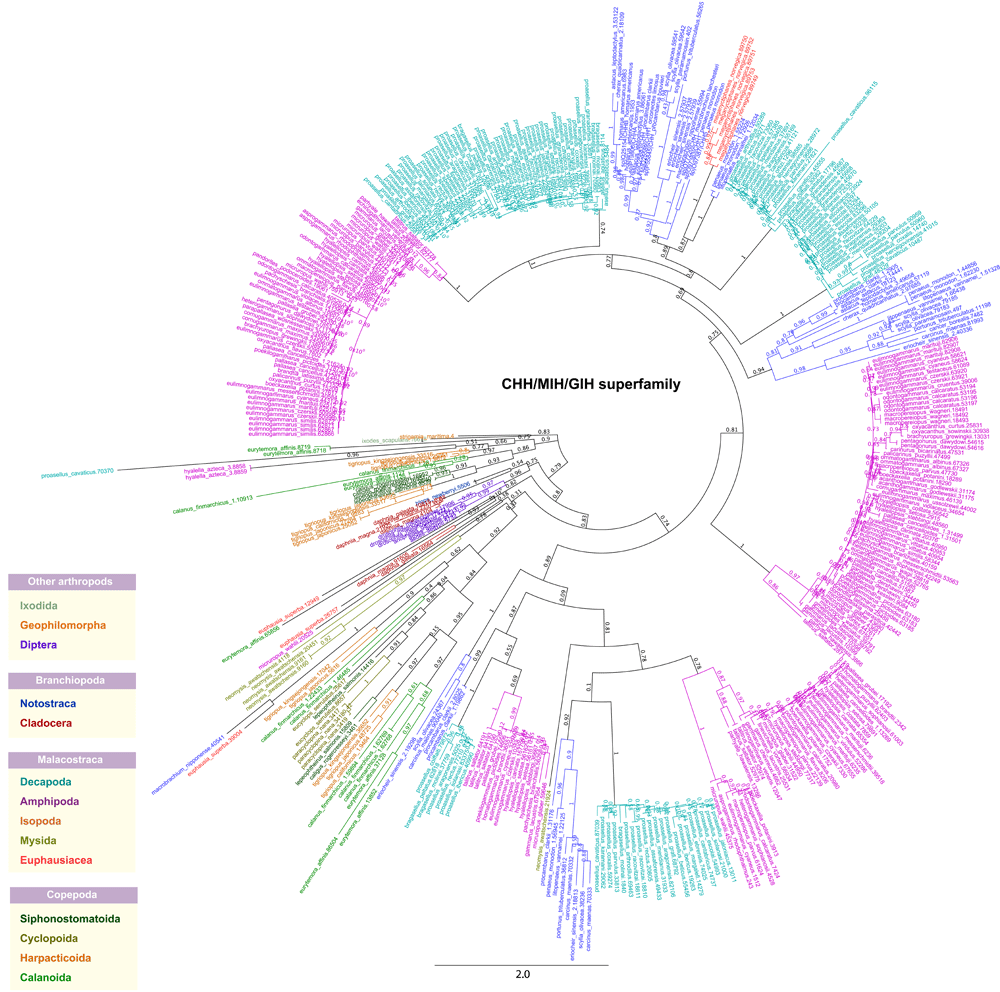
The tree was constructed using the maximum-likelihood method from an amino acid multiple sequence alignment. The node labels of each taxon are marked with distinctive colors denoted in the figure inset. Bootstrap support values (n=1000) are denoted as branch labels.
We have generated a high confidence list of CHH/MIH/GIH sequences from distantly related crustaceans. As a fundamental step in a broader endeavor this data is now available to the wider community to allow detail functional analyses pertinent to the next stages of neuropeptide research. Given the paucity of CHH sequences beyond decapod crustaceans, our analysis forms a promising basis for studies ranging from biochemistry to the evolution of this elusive superfamily.
Data supporting the conclusions of this study are provided as Supplementary Material and Dataset 1 and Dataset 2.
Dataset 1: Fasta file for CHH/MIH/GIH sequences in crustaceans and other arthropods. 10.5256/f1000research.13732.d19119428
Dataset 2: List of Pfam annotated CHH/MIH/GIH genes and associated e-values in crustaceans and other arthropods. 10.5256/f1000research.13732.d19119529
This work was supported by the EMBO Fellowship and the Human Frontier Science Program Fellowship to AGL.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Supplementary Table 1: List of accession numbers for species used in this study.
Click here to access the data.
Supplementary Table 2: Protein distance matrix (% identity) constructed from a multiple sequence alignment of CHH/MIH/GIH sequences.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
References
1. Chang W, Lai A: Mixed evolutionary origins of endogenous biomass-depolymerizing enzymes in animals. BMC Genomics. 2018; 19 (1). Publisher Full TextCompeting Interests: No competing interests were disclosed.
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Endocrinology
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Partly
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
References
1. Ohira T: Crustacean Hyperglycemic Hormone. 2016. 403-1 Publisher Full TextCompeting Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |||
|---|---|---|---|
| 1 | 2 | 3 | |
|
Version 1 23 Jan 18 |
read | read | read |
Click here to access the data.
Spreadsheet data files may not format correctly if your computer is using different default delimiters (symbols used to separate values into separate cells) - a spreadsheet created in one region is sometimes misinterpreted by computers in other regions. You can change the regional settings on your computer so that the spreadsheet can be interpreted correctly.
Click here to access the data.
Spreadsheet data files may not format correctly if your computer is using different default delimiters (symbols used to separate values into separate cells) - a spreadsheet created in one region is sometimes misinterpreted by computers in other regions. You can change the regional settings on your computer so that the spreadsheet can be interpreted correctly.
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)