Keywords
Pseudomonas aeruginosa, 16S rRNA, Alignment, PCR, phylogenetic tree, Blast, Sudan.
Pseudomonas aeruginosa, 16S rRNA, Alignment, PCR, phylogenetic tree, Blast, Sudan.
Pseudomonas aeruginosa is a gram-negative bacterium that is found widely in the environment and engages in various forms of interactions with eukaryotic host organisms. It is an opportunistic pathogen that is widely spread in humans, giving rise to a broad spectrum of infections in community and healthcare facilities1,2. Due to the extended spread of P. aeruginosa habitat, the control of the organism in a hospital setting is very difficult, and makes it practically impossible to prevent contamination3. The major threat is the infection of patients who are immunocompromised or those in burns, neonatal and cancer wards4,5. Infection of P. aeruginosa is still one of the main causes of death among the critically ill and patients with impaired immune systems in spite of the development of newer and stronger antibiotics.
Sequencing of 16S rRNA worldwide has provided interesting and useful information6–9. For instance, with the use of 16S rDNA sequencing, 215 novel bacterial species, 29 of which belong to novel genera, have been discovered from human specimens in the past 7 years of the 21st century (2001–2007). One hundred of the 215 novel species, 15 belonging to novel genera, have been found in four or more subjects10. In Sudan, there is deficient data on sequencing about bacteria; therefore it is important to investigate what kinds of bacteria affect Sudanese people. Consequently, the objective of this study was to isolate and characterize P. aeruginosa from samples obtained from Sudanese patients by sequencing the 16SrRNA gene.
This was cross sectional laboratory based study, conducted in Khartoum state in the period from January to April 2016. The study was approved by the Ethical and Scientific Committee of the Medicinal and Aromatic Plants and Traditional Medicine Research Institute, National Center of Research, Khartoum, Sudan (approval number 03-16) which ensures that all ethical considerations for conducting the research in a way that protects patient’s confidentiality and privacy are followed. Informed consent was obtained from the hospital laboratories (laboratory manager) after providing them with the ethical approval to collect samples during routine procedures from the microbiology laboratories. Participants’ privacy and confidentially was protected for all samples; personal information was not of great value in the current study and was thus not taken. Consequently, the Ethical and Scientific Committee waived the need for patient consent.
A total of 40 isolates of P. aeruginosa (all samples available) were obtained from three hospitals in Khartoum State (Al Ribat Hospital, Bahri Hospital and Souba Hospital).
The bacterial isolates were collected from sputum culture, urine culture and wound infection because these types of cultures were dominant. Standard biochemical tests7 were performed on all samples for the isolation and identification of the bacterial isolates and were performed at the Medicinal and Aromatic Plants and Traditional Medicine Research Institute (MAPTMRI), Department of Microbiology. P. aeruginosa presence was confirmed in all 40 samples. All media required for biochemical tests were obtained from LAB M, UK and Laboratories Flow Media, Sudan.
Bacterial DNA was isolated by the Chelex-based protocol7 for all samples without deviation from the methodology.
All bacterial genomic DNA were used as templates for PCR amplification of the 16S rRNA gene. The two primers used were 27F (5ʹ AGAGTTTGATCCTGGCTCAG-3ʹ) and 1495R (5ʹ CTACGGCTACCTTGTTACGA- 3ʹ) for forward primer and reverse primer, respectively (Macrogen, South Korea). The 25μL PCR reaction mixture (Intro Biotechnology, South Korea) contained 1μL DNA, 1x reaction buffer (10x) with 3mM MgCl2, 2.5U i-TaqTM DNA polymerase (5 U/μL), 2.5mM dNTPs, 1μL of 10 pmol of each primer, and 1x of gel loading buffer, followed by completing the volume to 25μL with nuclease free water. Thermal cycling conditions were as follows: 94°C for 5 min; 30 cycles of denaturation (94°C for 1 min), annealing (58°C for 1 min), extension (72°C for 2 min); final extension at 72°C for 10 min. PCR was performed on a Bio-Rad (DNA engine/Dyad Peltier) automatic thermal cycler. Duplicate PCR of every sample were carried out for confirmation11,12. PCR products (5μl) were analyzed by gel electrophoresis in 1.0% agarose stained with ethidium bromide. The results were photographed under ultraviolet light machine (Transillumnator; Uvite, UK) to detect the specific amplified product by comparing it with 100 base pairs standard DNA ladder (Figure 1) and the remains from PCR products were store at -20°C until sequencing.
Isolates were packaged according to the International Air Transport Association guidelines and shipped with authorized permission to Macrogen Company (Seoul, South Korea). Purification and standard forward sequencing of 16S rRNA were done by ABI Genetic Analyzer (Applied Biosystems). We randomly selected 20 isolates only for DNA sequencing due to the limitation of resources.
The chromatogram sequences were visualized using Finch TV program version 1.4.013. The nucleotide sequences of the 16S rRNA gene were searched for sequences similarity using online BLASTn14. Highly similar sequences (accession numbers KX108935.1, KT943978.1, JN609194.1, KX214108.1, KU672378.1, KU764451.1 and FJ648815.1) were retrieved from NCBI GenBank and subjected to multiple sequence alignment using BioEdit software version 7.2.5. Newick format was withdrawn from ClustalW12, to create phylogenetic trees in Phylogeny.fr software15. MEGA6 software version 0.06 was used for confirmation of phylogentic trees16.
A total of 40 clinical isolates were identified as P. aeruginosa by conventional methods, including growth characteristics, colony morphology, and biochemical tests. The results revealed that P. aeruginosa were dominant in wound infection cultures (42.5%), followed by (32.5%) from urine cultures and (25%) from sputum cultures.
The 16SrRNA in the isolates was amplified by PCR. In total, 20 were sent for characterization by sequencing of PCR products, 5 out of these 20 had a clear chromatogram, leading to further sequence analysis. Sequence analysis by BLASTn revealed 100% identity with P. aeruginosa from Iran (KX108935.1), China (KT943978.1), Australia (JN609194.1), Austria (KX214108.1), India (KU672378.1) and South Africa (KU764451.1), and slightly different from a sample from Australia (FJ648815): there was one substitution of A to G in position 148 (Figure 2). The phylogenetic tree of the 16S rRNA gene and other 16S rRNA genes obtained from the database revealed that the tree is classified into two branches. All isolates have a common ancestor. The isolate 120 is at the upper branch, isolate 113 and isolate 114 are sister groups, as shown in Figure 3.
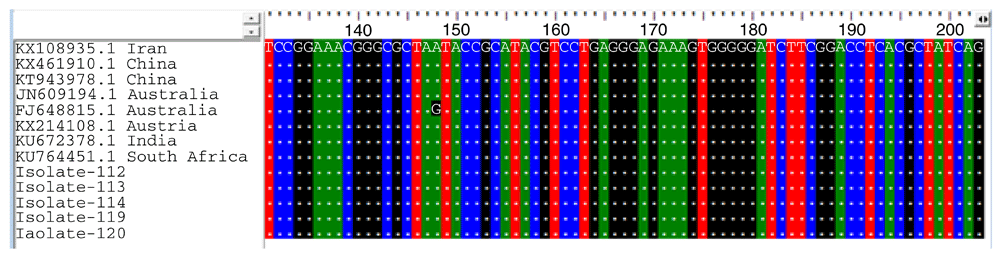
The isolates had 100% identify with samples from Iran, China, Austria, India, and South Africa. There was one substitution of A to G in position 148 from a sample from Australia (FJ648815).
P. aeruginosa can cause various clinical infections. In the present study, 42.5% of isolates were identified from wound infection. Beside the highly preserved conserved primer binding sites, 16S rRNA gene sequences include hypervariable regions with high conservation that have the ability to characterize species-specific signature sequences beneficial to the characterization of bacteria17,18. The present results agree with Didelot et al., who reported that 16S rRNA gene sequencing is now common in medical microbiology as a quick and inexpensive alternative to phenotypic approaches of bacterial identification19. We investigated the phylogenetic affiliation of the pseudomonads and reexamined the 16S rRNA sequence data available in public databases. According to an alignment of 16S rRNA sequences, we identified P. aeruginosa-specific signature sequences.
16S rRNA sequencing can be used to identify genetically atypical P. aeruginosa isolates from different origins. Our observation of 40 isolates from 40 samples, further showed the utility of 16s rRNA PCR amplification. This reveals the high specify of the primers and accuracy of the PCR. Sequencing of the 16S (rRNA) gene was used as an effective tool for determining phylogenetic relationships between bacteria. The features of this molecular target make it valuable and useful for bacterial detection and identification in the clinical laboratory.
In conclusion, 16S rRNA-based PCR assay and sequencing is highly specific, sensitive and reliable for identification of P. aeruginosa and its differentiation from other genotypic closely related Pseudomonas species. DNA sequencing of the 16S rRNA gene has been used as an effective tool to study bacterial phylogeny and taxonomy relationships between bacteria and for bacterial detection.
The results of the nucleotide sequences of the 16S rRNA gene were submitted in the GenBank database. Accession numbers: KX650648, KX650649, KX650650, KX650651 and KX650652.
The authors are grateful to Africa City of Technology, Medicinal and Aromatic Plants and Traditional Medicine Research Institute (MAPTMRI), Microbiology and Parasitology department, National Center for Research, Khartoum, Sudan and department Epidemiology, Molecular Epidemiology Laboratory Biology, Tropical Medicine Research Institute, National Centre for Research, Khartoum, Sudan for their kind support.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
No
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Not applicable
Are all the source data underlying the results available to ensure full reproducibility?
Partly
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Molecular Microbiology. Peptidoglycan characterization, Antibiotic resistance, betalactamases
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
I cannot comment. A qualified statistician is required.
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 25 Jul 18 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)