Keywords
plastome, Psidium, Galapagos, guayabillo
plastome, Psidium, Galapagos, guayabillo
Over a quarter of all vascular plant species are endemic to islands, making them hotspots of plant diversity and conservation (Kreft et al., 2008). In the Galápagos Islands, there are roughly 560 native species of plants of which approximately 32% are endemic (Lawesson et al., 1987). However, many of these endemic species have remained relatively unstudied since they were originally given scientific descriptions, making the study of the evolutionary histories of these unique taxa difficult. In the present study, we constructed the complete plastome sequences of two species of Psidium (guava) from the Galápagos Islands, one endemic and one currently unidentified in hopes of facilitating future work on the evolutionary relationships of these species.
This research is authorized under the permit: MAE-DNB-CM-2016-004 in compliance with Ecuadorian regulations.
Leaf samples were collected during May of 2017 from the Galápagos endemic Psidium galapageium Hook (commonly known as guayabillo) on the island of San Cristobal (0.89094°S, 89.43769°W) and from an unidentified Psidium species on the island of Santa Cruz (0.62313°S, 90.38581°W). Based on morphological similarity, the Psidium sp. individual is suspected to be P. acidum (Landrum, 2016), but no reference or barcode sequence from P. acidum is available for confirmation.
Leaf tissue was desiccated immediately after harvesting using silica gel. DNA extractions were performed using a Qiagen DNeasy Plant mini kit (Qiagen, Inc.). Sequence data was generated in the form of paired-end, 150 bp reads using a KAPA library prep kit (Roche Sequencing) and sequenced on an Illumina HiSeq 4000 platform (Illumina, Inc.).
Reads were quality and adapter trimmed using Trim Galore! version 0.4.3 with a minimum phred score value of 20 and minimum read length of 50 bp. Filtered reads were then aligned to the Psidium guajava plastome reference available at NCBI (Accession: KX364403) using the mem function within BWA version 0.7.15 (Li & Durbin, 2009). Consensus plastome sequences were generated using the mpileup function within samtools version 1.8 followed by the call and consensus functions within bcftools with a minimum depth of coverage of 10x (Li et al., 2009). Using IRscope (Amiryousefi et al., 2018), the P. galapageium and Psidium sp. plastomes respectively were confirmed to contain a large single copy of 88,268 bp and 87,747 bp and a small single copy of 18,465 bp and 18,490 bp separated by two inverted repeats of 26,071 bp and 26,360 bp for total lengths of 158,875 bp and 158,957 bp (Figure 1).
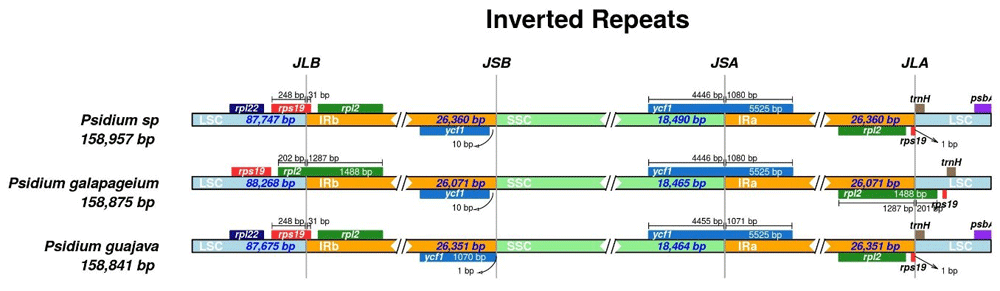
The circular genomes have been linearized for illustration.
Annotations were generated using the program Plann (Huang & Cronk, 2015). Of the 132 gene features annotated previously in the Psidium guajava (guava) chloroplast genome on NCBI (Accession: KX364403), all were recovered in the Psidium sp. and P. galapageium plastome sequences. The non-identity of the two taxa sampled is evidenced by the absolute pairwise sequence divergence of the concatenated sequences of three conserved genes (MatK, psbA, and rbcL) which have been successfully used as barcodes previously in Psidium (Kress et al., 2009). Sequences were aligned using MUSCLE within MEGA version 7.0.26 (Tamura et al., 2007), and the number of nucleotide differences were counted between these alignments to estimate divergence. A total of 35 differences were observed among 4011 sites (0.87% uncorrected divergence) between P. guajava (Accession: KX364403) and P. galapageium, 45 differences (1.1%) between P. guajava and Psidium sp., and 40 differences (0.99%) between P. galapageium and Psidium sp.
Voucher specimens for P. galapageium and Psidium sp. are available at the Charles Darwin Research Station herbarium (Index Herbariorum code CDS) with accession numbers 3053515 and 3053562, respectively. The corresponding plastome sequences for P. galapageium and Psidium sp. are available at NCBI with accession numbers MH491846 and MH491847, respectively.
This work was supported by a Louise Coker Fellowship from the University of North Carolina at Chapel Hill (UNC).
All funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The authors would like to thank the staff of the Galápagos Science Center, Galápagos National Park, Heinke Jäger and Diana Flores (Charles Darwin Foundation), and the High Throughput Sequencing Facility at UNC for assistance with fieldwork, curation, and sequencing. Special thanks to Marcelo Loyola for the invaluable help during the fieldwork.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the rationale for creating the dataset(s) clearly described?
Yes
Are the protocols appropriate and is the work technically sound?
Yes
Are sufficient details of methods and materials provided to allow replication by others?
Partly
Are the datasets clearly presented in a useable and accessible format?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Myrtaceae Systematics, Plant Phylogenetics
Is the rationale for creating the dataset(s) clearly described?
Yes
Are the protocols appropriate and is the work technically sound?
Yes
Are sufficient details of methods and materials provided to allow replication by others?
Yes
Are the datasets clearly presented in a useable and accessible format?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Vascular plant systematist
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 30 Aug 18 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)