Keywords
Measles virus, Phylogeny, Clade A, Nigeria
Measles virus, Phylogeny, Clade A, Nigeria
Measles is an acute viral illness characterised by a prodromal illness of fever, coryza, cough, conjunctivitis and presence of Koplik spots followed by the appearance of a generalised maculopapular rash. The illness is either mild or severe depending on the immune and nutritional status of the infected person. It is caused by measles virus (MV), which is a negative sense single stranded RNA virus of the family Paramyxoviridae, genus Morbillivirus1. The virus is relatively antigenically stable and monotypic but has genetic variation in the hemagglutinin (H) and nucleoprotein (N) genes2. Measles is one of the leading causes of death among young children despite the availability of a safe and cost effective vaccine for over 40 years3. It accounted for over 145700 deaths in 2013 globally, 51% of which are from World Health Organization (WHO) defined African region4. Nigeria still ranks high among countries reporting measles infection yearly, with over 7000 suspected and over 3000 confirmed cases reported in 20144.
The 450 nucleotides that code for the COOH-terminus of the N protein is the single most variable part of the measles genome and analysis of this part of MV isolates or clinical specimens has helped to classify measles into eight clades (Clades A-H) and 23 genotypes5. Different measles genotypes and strains circulate in specific geographical regions5,6. Measles virus clades B3, D4, D5, D8 and H1 were isolated in the WHO Americas region between 2007 and 2009, genotypes D6 and D9, in addition to those found in the Americas, are also found in the European region, and genotype B3 is the major circulating strain in the Africa region in this period6.
Previous reports on circulating measles strains from Nigeria have indicated the circulation of only genotypes B3 cluster 1 and 2. However, due to the absence of integrated molecular surveillance in measles’ elimination programs, there has been only a few sequence data of Nigerian measles isolates, especially in recent times7,8. Molecular surveillance is essential in order to observe the changes in viral genotypes over time in a particular region. It is also an important tool in assessing the effectiveness of vaccination programs9. This study reports the molecular characterization of a previously unreported genotype of measles virus from Nigeria.
As part of the standard care routine, a nasopharyngeal swab was collected from a 3-year-old child of suspected measles infection presenting with fever, maculopapular rash, cough and conjunctivitis at the Oni Memorial Children’s Hospital in Ibadan, Oyo State, Nigeria, in November 2010. The sample was transported to the lab in virus transport medium under reverse cold chain. The swab was inoculated into tissue culture flask of VeroSLAM cell line and observed for cytopathic signs for seven days.
A nasopharyngeal swab was used as the sample type because it is the recommended site by the United States Centers for Disease Control and Prevention. In addition, measles virus is shed up to 10 day after the onset of rash and after the resolution of vireamia increasing the chances of viral nucleic acid detection up to the 14 day after the onset of rash.
Viral RNA was extracted from the throat swab collected and supernatant from the cell culture using QIAamp® Viral RNA kit by QIAGEN Valencia USA, according to the manufacturer’s instruction.
Extracted RNA was synthesised to cDNA by reverse transcription using a commercial kit (SCRIPT DNA synthesis kit by Jena Bioscience® GmbH, Germany). Nested PCR was performed using two sets of primers: First round, fwd MN5 5-GCCATGGGAGTAGGATGGAAC-3; rev MN6 5-CTGGCGGGCTGTGTGTGGACCTG-3 and nested inner primers Nfla 5-CGGGCAAGAGATGGTAAGGAGGTCAG-3, Nr7a 5-AGGGTAGGCGGATGTTGGTTCTGG-3, as previously described by Kremer et al.8 using Applied Biosystems GeneAmp PCR System 9700 thermal cycler. Cycling conditions for both first and second reactions is as follows: 35 cycles of 94°C for 30 seconds, 55°C for 1 minute, 72°C for 1 min preceded by 94°C for 5 minutes and a final elongation at 72°C for 5 minutes8.
Purified amplicons were sequenced at Jena Bioscience Laboratory (Germany) by Sanger sequencing method using the second round primer. Sequence data obtained was edited and assembled with Bioedit software version 7.0.5, sequence similarity was determined by Basic Local Alignment (BLAST). The query sequence was aligned with reference sequences downloaded from GenBank with the help of Measles Nucleotide Surveillance (MeaNS) database. Table 1 shows the names and GeneBank accession numbers of the reference sequences used for the analysis. The basis of sequence selection was predicated on selection of characterized isolates reported in publications from WHO European region laboratory7,8 using CLUSTALW software. Phylogenetic trees were constructed in Mega version 6.06 software using Maximum likelihood and Neighbor joining methods with p distance model and 1000 bootstrap replicates.
Both the swab and tissue culture sample showed the expected 560 base pairs after nested RT-PCR. Phylogenetic analysis of the measles sample MViIbadan/NIE/11.10 sequence (ccession no LN876569.1) gave a distinct BLAST search result (Supplementary File 1). From the search it was observed that the sequence with ascension no JF727650.1 Leningrad is the most closely related virus sequence with our Ibadan strain. The phylogenetic tree generated revealed that the Ibadan prototype Clade A strain co-clustered with sequences close to the vaccine Edmonston Zagreb stain with 98% bootstrap value (Figure 1), suggesting the possibility of common ancestral origin. Molecular Epidemiology of Measles virus in Nigeria classifies isolates into two distinct groups within genotype B3, B3 cluster 1 and B3 cluster 27,8. The B3 genotype was assigned after exhaustive genetic analysis of MV strains from Lagos and Ibadan in 19998.
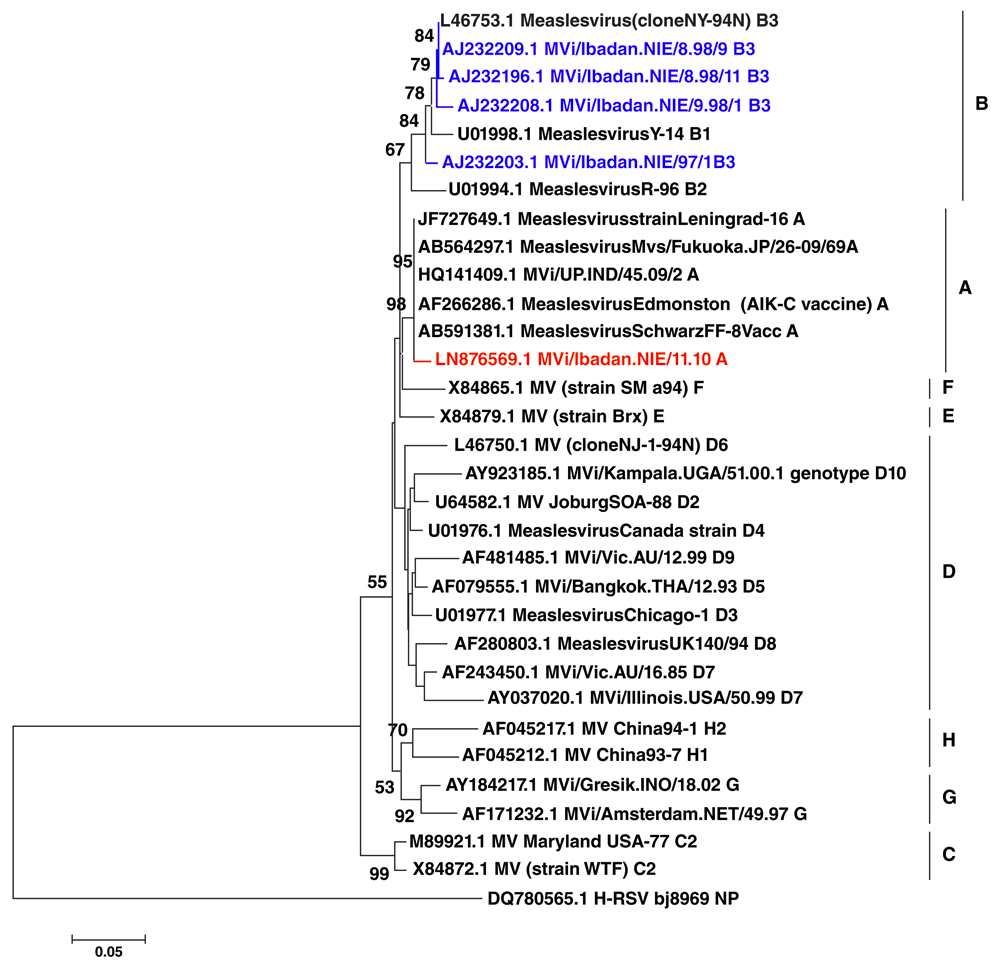
The study isolate is shown in bold red font; clinical strains from Nigeria are shown in blue font. Clades are indicated beside the black horizontal lines. The GenBank accession numbers are indicated first in the sequence labels, bootstrap values are indicated if ≥ 50%, phylogenetic tree was constructed using the Neighbor joining algorithm in MEGA 6.0 with 1,000 bootstrap replicates.
Our current study describes isolation of a different strain of measles from the previously described genotypes (MViIbadan/NIE/11.10). This virus was recovered from the throat swab of a 3 year old child with no documented history of vaccination at Oni Memorial Children Hospital in Ibadan Nigeria in November 2010.
Measles virus of genotype A has previously been detected in acute cases of measles in South and North America, China, Japan, Eastern Europe, Finland and the UK and South Africa over the last 40 years10, but there had not been any report of Clade A virus in West Africa. The virus isolated in this study is from a child who had never received any form of measles vaccination and there was no activities involving measles virus vaccine in the laboratory as at the time of virus isolation. Molecular epidemiologic information has revealed circulation of several measles genotype in Africa in recent times. Clade B viruses is reported to be endemic in central and western parts of sub-Saharan Africa while genotypes D2 and D4 has been continually detected southern and eastern parts of Africa and genotype C2 in northern Africa11–14 but no virus of Clade A has been reported in Africa recently.
In this study, although we did not independently investigate the immediate or remote factors that could have led to the introduction/emergence of this virus genotype in Nigeria, we postulate that the virus could have been probably imported from another continent or could have been an undetected circulating strain. This is because active molecular surveillance is not in place in Nigeria, and the last report of MV molecular epidemiology was in 20058. This leaves a huge gap in research information and multiple introductions of diverse measles genotypes might have taken place undetected over time. We therefore advocate for the establishment of an institution of a national surveillance program, and the inclusion of molecular epidemiology alongside national and global authorities, such as the WHO and Federal Ministry of Health Nigeria, to help achieve measles elimination goals in the country and globally.
The sequence of the identified isolate has been deposited in GenBank under the accession number LN876569.1.
Ethical approval was obtained from Oyo State, Hospital Management Board (Ibadan, Nigeria), approval number OY/HMB/REC-140010, to conduct this study. The parent of the infected child reported in this study also provided written informed consent for sequencing of the sample and use in research before the sample was collected.
The authors would like to acknowledge the contributions of Prof D.O Olaleye and Dr G.N Odiabo during the course of this work. The effort of Mr B.A Olusola is also appreciated during the course of the field work. A.O. Faneye was a PhD student during the time of this study and this report is part of her PhD research project.
Supplementary File 1: Blast search result for MViIbadan/NIE/11.10 Genbank no LN876569.1.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
References
1. Roy F, Mendoza L, Hiebert J, McNall RJ, et al.: Rapid Identification of Measles Virus Vaccine Genotype by Real-Time PCR.J Clin Microbiol. 55 (3): 735-743 PubMed Abstract | Publisher Full TextCompeting Interests: No competing interests were disclosed.
Reviewer Expertise: Virology (RNA viruses)
Competing Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 07 Feb 18 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)