Keywords
genome, exposome, interactome, exposure, wearable, health tracker, fitness device, fitness tracker, sleep, heart rate
genome, exposome, interactome, exposure, wearable, health tracker, fitness device, fitness tracker, sleep, heart rate
Human health and performance is understood to be affected by both nature (genome) and nurture (activities & environment). One notable example of the combined effects of genetics and the environment on health is the identification that the GRIN2A gene significantly modulates risk for developing Parkinson’s disease, but only in heavy coffee-drinkers1. This study provides proof that inclusion of quantitative measures of environmental factors can help identify important genes that would be otherwise missed in GWAS studies that ignore exposures. However, the challenges associated with designing and implementing broad quantitative studies of complex interactions at scales sufficient to achieve sufficient statistical power are considerable.
There are multiple efforts underway that are making progress toward addressing the challenges of integrating genome, interactome, and exposome2 data to support focused scientific studies. The Institute of Systems Biology’s Hundred Person Wellness Project3 and 100K Project4 are integrating genomics, monitors, and blood sampling to build on the pioneering N-of-one work conducted by Larry Smarr5 and Michael Snyder6,7 to articulate the vision and promise of predictive, preventative, personalized, and participatory (P4) medicine8. Orion Bionetworks9 is combining traits, genetics, and interactome with a focus on brain disorders. Sanchez et al.10 has also proposed exposome informatics integrating the genome, phenome, and exposome. Systems integrating personal sensors and exposome have been developed by Doherty & Oh11 and Nieuwenhuijsen et al.12. Other relevant available resources include PhysioNet13 and MOPED14. The Human Longevity project15 is examining genome, microbiomes, and metabolites of volunteers. Lifestyle affects human microbiomes16,17. While these projects all share the common elements of longitudinal integration of heterogeneous biomedically relevant data, each either focuses on a relatively narrow set of measurements or relies on custom data storage and analysis architectures that do not provide a scalable foundation for larger-scale integration across studies to enable meta-analysis of data from multiple studies.
The Integrated Biomedical System is being developed as an open source platform for integrating genome, interactome, and exposome data that provides a unifying model to promote more open data sharing and analysis. The software architecture with multi-scale operability design intended to scale from running on a single laptop/workstation as a standalone system with an embedded private local database, to a study platform, to large-scale implementations all using standard scalable web technology stacks.
The Integrated Biomedical Project description and written consent form (Protocol # 1312006029) was reviewed and approved by the Massachusetts Institute of Technology (MIT) Committee on the Use of Humans as Experimental Subjects (COUHES) for the initial 20 volunteers and the expansion to 40 volunteers. COUHES is the MIT Institutional Review Board (IRB). This project used no recruitment. All volunteers learned about the project from other volunteers, typically by expressing interest in the devices being worn. Upon expressing interest to the principal investigator, the project was fully explained and a written consent form was provided and the project explained with multiple voluntary options. The project principal investigator and co-investigator were primary points of contacts for all volunteers. All volunteers signed the written consent form approved by the MIT COUHES and provided their signed form to the project principal investigator. Volunteers have full choice of all elements of the research project that they elect to participate in or not. Volunteers may elect to have all or any subset of their data removed from the system at any time. Volunteers either elect to opt-in or opt-out of notification of any possible data abnormalities detected.
Extract, transform, and load (ETL) modules were developed for 23andMe SNPs18 files, SwissProt19 dat file, DrugBank20 XML file, NCBI Gene21 gene file, PharmGKB pathways22, and Protein Data Bank (PDB) protein structures23. After SwissProt sequences and PDB protein structures were loaded, the structure coordinates were mapped to sequence residues with the included lib/utilities/align_pdb.rb tool; this enables the visualization of residues and variants on structures. Interface modules were developed to allow individual or pooled variants to be visualized on protein structures with the integrated Jmol24 structure viewer.
Interactome data included in the pilot collection described herein includes heart rate, interbeat interval (IBI), and electrocardiogram (ECG), skin temperature, skin conductance, galvanic skin response, and respiratory rate. These aggregated data were collected by a diverse collection of commercially available wearable physiological monitoring devices. All volunteers were offered a Basis B1 watch25 and Polar Loop H7 heart rate monitor26. A subset of volunteers are evaluating Hildago Equivital EQ-02-SEM27, Empatica E328, Mio Link29, and Zephyr BioHarness 330 devices. Data logging functionality was not built in for the Polar Loop and Mio Link heart rate monitors, so these data streams were wirelessly synced and stored continuously on co-worn Actigraph Actisleep device. ETL modules were developed for Basis B1 json files31, Actigraph heart rate csv or dat files (including Polar Loop and Mio Link), Empatica E3 zip files, Hidalgo Equivital SEM2 persisted summary csv files, Zephyr BioHarness summary csv files, vocal recordings and associated Matlab .mat files. Data displays include Ruby gems and JavaScript plugins: Google Maps32, jQuery33, lazy_high_charts34, Highstocks35, Data-Drive Documents (D3)36, FullCalendar37, rails3-jquery-autocomplete38, and more. The graphical user interface for “Data Loading” provides the ability to download data from the Basis web site and drag and drop interfaces for easy file uploads for each of the device ETL modules.
Activity and sleep were monitored continuously using wearable and personal electronic devices that used algorithms to process raw data provided by built-in 3-axis accelerometers. Data describing daily nutrition, prescriptions, and over-the-counter medications were collected manually and provided by a subset of volunteers. Devices used by volunteers for continuous data collection included the Fitbit Flex, the Basis B1 watch, Actigraph ActiSleep monitors, basic Actigraph activity monitors GT3X+, Jawbone Up, and smart-phone apps including MyTracks, and Sleep Cycle. ETL modules were developed for Fitbit csv files, Jawbone csv files, Actigraph39 sleep csv files, MyTracks app40 csv files, and Sleep Cycle app41 csv files. Additionally to demonstrate the ability to integrate other publicly available data, modules were developed for integration of EPA AirData (daily and hourly csv files42), and foods43. Graphical user interfaces were developed for entering activities, events, meals, drinks, prescriptions, and over-the-counter medicines. Multiple volunteers submitted oral swab samples for metagenomics sequence analysis when sick (cued data collection).
The Integrated Biomedical System was developed on the Ruby on Rails44 platform with Ruby gems and JavaScript plugins. The Rails platform supports multiple SQL relational databases including MySQL and no SQL databases such as Mongo DB. MySQL, Oracle, Mongo DB, etc. all scale to over a billion records in a single table. The underlying architecture and approach can be extended to handle a variety of additional data sources. To facilitate data exchange between sites, global unique identifiers (guids) are used. The Integrated Biomedical System and Rails can be installed on computers ranging from stand-alone on a laptop/desktop to servers running Windows OS, Mac OS, Linux, or Unix. Individuals can install and run this system for personal use without needing to set up a web service; to facilitate this the default configuration uses the Sqlite3 database, which installs with the Rails setup. Switching to MySQL or Oracle requires database software installation and a 5-line update to the Rails database.yml configuration file with updated database instance details. To facilitate bulk loading of large numbers of data files, command line interfaces for each ETL module are included in the app/utilities folder.
The Integrated Biomedical System (version 1.0) is developed as a Ruby on Rails (versions 3 & 4) application. Current JavaScript libraries and versions are included in the Ruby on Rails Gemfile with the inclusion of JQuery, D3, FullCalendar, Highcharts, JSmol24 PDB structure viewer, and more. The Integrated Biomedical System can be optionally configured as a web site with Apache httpd web server plus Passenger (Phusion). The database schema is available in a MySQL Workbench schema in the docs folder for the application. The Integrated Biomedical System has been tested with both Sqlite3 and MySQL relational databases; it should work with most if not all Rails supported databases. The application Readme and GitHub site (https://github.com/doricke/ibio) list the 10 standard Rails application setup steps to setup this Rails application. Initial user accounts can be configured in the db/migrate/20131217194515_create_individuals.rb file.
The Integrated Biomedical System can be run as a local application with the “rails server” command and a web browser for http://localhost:3000/ or configured to run as a web application with Apache httpd server. The graphical user interface navigation control panel is a set of eight ovals containing text and image links to interfaces within the application, see top of Figure 1. Users can upload data through the web interface (Figure S1). A set of command line utilities are included for administrator loading of data (Table S1).
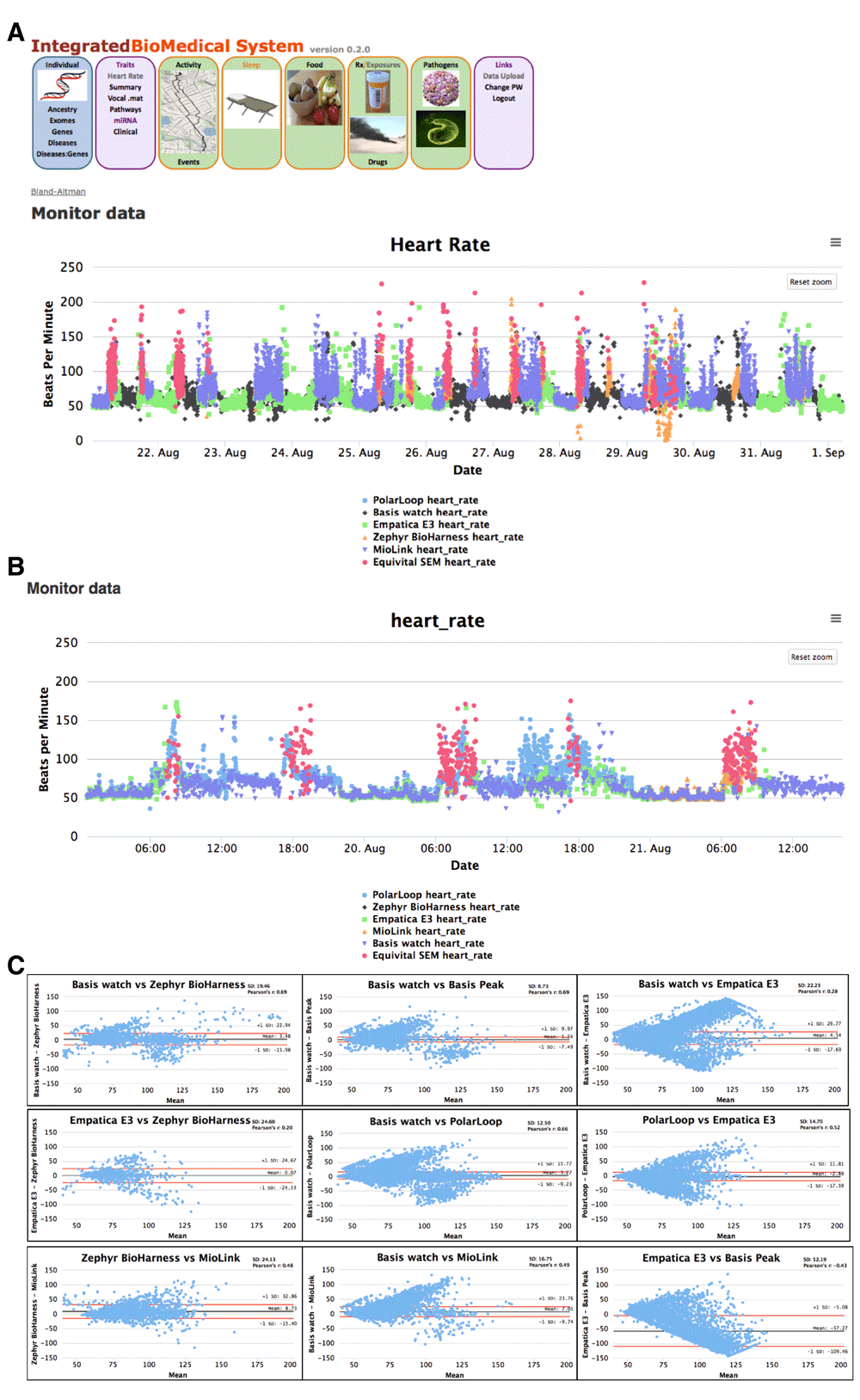
(A) Screen shot of heart rate beats per minute measurements for a volunteer wearing Basis B1 watch, Empatica E3, Zephyr BioHarness, Hildago Equivital SEM2, and Mio Link devices. SEM2 values were filtered for minimum quality values of 70 with selection of median value; (B) Zoomed in view of heart rates illustrating measurements at different activity levels; and (c) Bland-Altman plots comparing measurements from the heart rate tracking devices with corresponding Pearson r correlation values.
Heart rate monitoring. Heart rate monitoring devices provide heart rate, interbeat interval (IBI), and electrocardiogram (ECG) measurements. Heart rate measurements for multiple devices for an individual are shown in Figure 1. Hidalgo Equivital SEM2 and Zephyr BioHarness were typically worn only during more active periods. Lower Zephyr heart rate values observed on Aug. 29 likely resulted from the contact pads drying out during a period of extended wearing with low activity level. Some data gaps result from the need for device battery recharging (Empatica E3 - daily and Mio Link every 8 to 10 hours). Higher correlations of results are observed for periods of sleeping and light activity. This observation is consistent with previous anecdotal observations of data accuracy and coverage decreases for many wearable sensors during periods of high activity.
Sleep monitoring. Multiple devices tested provide top-level estimates of nightly time asleep and number of sleep interruptions. Some devices also attempt to break down the sleep time into sleep phases (light, deep, and rapid eye movement - REM sleep). This data was integrated to enable comparisons of sleep classifications assigned by these devices (investigation of the accuracy of these estimates vs. gold-standard polysomnography was beyond the scope of the present work). Example longitudinal measurements from a single individual collecting data in parallel using Jawbone Up, Basis B1, Fitbit, and ActiSleep are shown in Figure 2. Analytical modules enabling pairwise comparisons of unfiltered nightly time asleep estimates between different devices were developed and integrated into the Integrated Biomedical System. Simple comparisons of daily total time asleep reported across the range of devices revealed a lack of correlation for most device pairs as measured by Pearson r statistics. Likewise, finer-grained estimates of light sleep (provided by Basis and Jawbone) and deep sleep (Jawbone) compared to deep sleep plus REM sleep (Basis) were also poorly correlated. Only the two Actigraph algorithms, Sadeh and Cole-Kripke, which were run on the same raw Actigraph sensor data produced highly correlated results (r of 0.97).
Global Position System (GPS) tracking of outside activities available in the Integrated Biomedical System from smartphone or GPS data can provide continuous localization for an individual. This data enables a range of potentially useful correlations to be determined including correlations with data from nearby EPA or other air quality monitoring station(s) as an initial step toward quantitative tracking of individual exposures. Inferred exposure levels can be estimated from nearby sensors for a wide variety of measured pollutants, particulates42, and pollen levels45. Figure 3 illustrates NO2, PM2.5, carbon monoxide, and ozone exposures for an afternoon walk.
Genome, interactome, and exposome all influence an individual’s wellness. The Integrated Biomedical System was developed to demonstrate the ability to begin integrating these heterogeneous data sources in near real-time for individuals. This was accomplished using an architecture that can operate on a stand-alone laptop or desktop personal computer (PC) to provide additional privacy and security and can be connected seamlessly to voluntarily transfer selected data to centralized highly scalable systems built on the same data architecture that can integrate data from many thousands or even millions of individuals. This approach could provide a path to developing new crowd-sourced models for large-scale prospective/retrospective studies of how individual combinations of genomic and environmental factors correlate with a range of human health and performance traits. Individual monitoring devices, genetic data, blood biochemistries, nutrition, exposures, illnesses, vocal and additional data have been organized and integrated into a unified system. Using the same tools and architectures, additional quantitative lab results and diagnostic data like images and physiological monitoring system data can be added to further increase the research scope of the system. Incorporation of additional natural language processing tools and data architecture modifications can enable text-based metadata collections (e.g. regular symptoms logging from personal health blogs, social interaction details from social media platforms, information from electronic health records) to be included in future versions of the system. Furthermore, these personal datasets can be combined with relevant public datasets and other non-public data to provide new insights into health-associated effects to support detailed N-of-1 and population retrospective analyses.
As large-scale DNA sequencing costs continue to decrease, sequencing an individual’s DNA becomes more affordable and practical. Current costs enable exome sequencing of individuals for less than $1,000. In a few years, the costs for whole genome sequencing for individuals is projected to be below $1,000 for very large studies. The quality and completeness of results can be estimated by coverage, but room for improvement is illustrated by the Proton and Illumina exome results correlated with 23andMe SNP profiles. While tools exist to characterize variants (Polyphen2, SIFT, etc.), the potential to correlate variants with protein structure/function, physiology, molecular biomarkers, etc. typically is done manually and within studies with a single focus. Integrating genomic data with interactome and exposome data will help create new opportunities for turning data into new discoveries and knowledge. The Integrated Biomedical System also supports detailed analysis of variant analysis for genes, proteins, pathways, individual SNPs, and other variant types. Future inclusion of raw genomic sequencing data and connections with a variety of genome viewers is straightforward using this extendable data and software architecture. As advances in DNA sequencing technology enable more widespread access to genomic data for individuals, the ability to correlate that data with quantitative interactome and exposome data will become increasingly important. Together, these data can broadly enable efforts to elucidate the interplay between genomic and environmental factors that contribute to complex individual human traits and health.
Cognitive performance and health phenotypes can be assessed through a variety of indirect methods including analysis of biomarkers in blood, psychomotor vigilance task (PVT), profile of mood states (POMS), automated neuropsychological assessment metrics (ANAM), speech analysis, facial and eye movement tracking, electroencephalography (EEG), and similar approaches. These assessments and others have been developed and used quantitatively define progressions of important traits/symptoms in individuals experiencing a number of conditions including depression46, posttraumatic stress disorder (PTSD), and traumatic brain injury (TBI), as well as environmental stressors including sleep disruption, etc. Data streams produced from these assessments combined with traditional measurements of traits, molecular biomarkers, and clinical data to provide a new platform for gaining insight into the underlying physiology individual health, fitness, and well-being. Retrospective analysis of large-scale collections will provide future biomedical discoveries. Increasing proportions of future biomedical discoveries will be driven by the ability to effectively collect, manage, and interpret massive amounts of heterogeneous data. Enhancements to integrate additional interactome data types and analysis tools are currently underway and these features will be included in future releases.
Asthma and COPD affect 18.7 and 6.8 million individuals in the United States47. Environmental exposures can exacerbate these conditions48. Asthma can be triggered by particulate matter, ozone, sulfur dioxide, nitrogen oxide, and pollens49. Devices, including smart phones, with GPS tracking ability enable the possibility of data integration with environmental monitoring data. Nearby monitoring stations and mobile monitoring devices provide weather and exposure estimates that can be correlated using time stamped GPS positional information. Monitoring stations track a rich variety of environmental exposure data42. While the current system provides incomplete coverage, it demonstrates a viable path to incorporation of additional sensor streams (including indoor air quality sensors, UV exposures, etc.) and activity-based estimates of indoor vs. outdoor exposures. It will be possible to provide increasingly complete individualized and integrated quantitative estimates of specific exposures that can be correlated with possible health effects, symptoms, and well-being. Larger and more complete data sets enabled by integrated systems like the one described here, can play a key enabling role for more quantitative genome vs. environment studies in the future.
The Integrated Biomedical System is being developed as an open source platform for individual health, fitness, and in the future wellness promotion. Data visualization, data mining, and new big data approaches will be integrated into the data analysis capabilities that will continue to expand over time. With the goal of creating an open data architecture that supports data exploitation and decision support, this system aims to provide useful information to individuals, medical personnel, researchers, and decision makers. Individuals can run this system on their home computer for use with their own data (and family members). This system will also support longitudinal studies integrating genome, interactome, and exposome heterogeneous data sources. Improving interfaces for user friendliness with valuable feedback and data visualization will be essential for user acceptance, continued use, and progress towards wellness promotion.
Latest source code: https://github.com/doricke/IBio.
Archived source code as at time of publication: https://doi.org/10.5281/zenodo.115633150
Software license: GNU General Public License, version 3.0.
The heart rate and sleep tracking data are included in Ricke, Darrell, 2017, “Integrated Biomedical System”, doi:10.7910/DVN/DEEHI251, Harvard Dataverse, V4.
The Equivital SEM data are included in Ricke, Darrell, 2018, "Integrated Biomedical System Equivital SEM", doi:10.7910/DVN/FD4B6C52, Harvard Dataverse, V1.
This work is sponsored by the Assistant Secretary of Defense for Research & Engineering under Air Force Contract #FA8721-05-C-002. Opinions, interpretations, recommendations and conclusions are those of the author and are not necessarily endorsed by the United States Government.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The authors would like to acknowledge Carl Ricke, Freelance Illustrator at Wanderbots, for graphic artwork.
Figure S1: Integrated Biomedical System web interface.
Click here to access the data.
Table S1: Integrated Biomedical System command line utilities for data loading. These extract, transform, and load (ETL) modules provide administrator tools capabilities for bulk loading of data files. Tools are run with the prefix “rails runner lib/utilities/<ETL loader.rb> <parameters>.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
No
Is the study design appropriate and is the work technically sound?
No
Are sufficient details of methods and analysis provided to allow replication by others?
No
If applicable, is the statistical analysis and its interpretation appropriate?
Partly
Are all the source data underlying the results available to ensure full reproducibility?
No
Are the conclusions drawn adequately supported by the results?
No
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Clinical research and IT infrastructures
Is the work clearly and accurately presented and does it cite the current literature?
No
Is the study design appropriate and is the work technically sound?
No
Are sufficient details of methods and analysis provided to allow replication by others?
Partly
If applicable, is the statistical analysis and its interpretation appropriate?
Not applicable
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
No
References
1. Kumar S, Abowd G, Abraham WT, al'Absi M, et al.: Center of Excellence for Mobile Sensor Data-to-Knowledge (MD2K).IEEE Pervasive Comput. 16 (2): 18-22 PubMed Abstract | Publisher Full TextCompeting Interests: No competing interests were disclosed.
Reviewer Expertise: Biomedical informatics, exposome informatics, translational bioinformatics
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 08 Feb 18 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)