Keywords
DNA Methylation, Gene Silencing, lncRNA, ncRNA, lincRNA, MORT, ZNF667-AS1, Epigenetics
DNA Methylation, Gene Silencing, lncRNA, ncRNA, lincRNA, MORT, ZNF667-AS1, Epigenetics
MORT was originally found as a transcript silenced during in vitro immortalization of human mammary epithelial cells1. Like a significant majority of lncRNAs, MORT’s molecular function remains enigmatic. The MORT gene is specific to higher primates, is expressed in all normal human cell types, and MORT RNA is located predominantly in the cytoplasm1. Analysis of MORT expression and the DNA methylation state of its promoter in 17 cancer types from The Cancer Genome Atlas (TCGA)2, which represent the 10 most frequent cancers in males and females, showed MORT is epigenetically silenced in 15 of 17 these cancers1. Based on the data from the original in vitro study1, we predicted epigenetic MORT silencing occurs early in human carcinogenesis and therefore could be seen in premalignant lesions, such as ductal carcinoma in situ of the breast and colonic adenomas. We used data from clinical samples from published genomic data sets3–8 to address this possibility, and indeed, MORT loss occurs prior to or at the stage of pre-malignancy and not thereafter9. Taken together these facts suggest that MORT transcript has a tumor suppressive role and is not simply an epigenetic “passenger error.”
Since our previous analysis of MORT in TCGA datasets was not exhaustive and only reported on 17 out of 33 TCGA cancer types, the goal of this short study was to extend our earlier work and complete the analysis of MORT DNA methylation associated gene silencing in the final 16 TCGA cancer types.
We integrated the MORT expression level and the DNA methylation state of its promoter region using TCGA data as described before1. The Illumina HiSeq RNA-seq and HumanMethylation450 DNA methylation data for samples of 16 TCGA cancer types listed in Table 1 were downloaded from the GDC data portal. The data were analyzed in the R programming environment, version 3.4.210. The mean RNA-Seq rpkm values for the two exons constituting the MORT RNA were plotted against the mean DNA methylation beta value of the 7 CpGs from the MORT promoter region for the individual samples of each cancer type. The Spearman correlation coefficient rho between the MORT RNA level and the DNA methylation of MORT promoter was calculated using the function cor.test.
The numbers of primary tumor and normal samples for which both the MORT RNA expression and the MORT promoter DNA methylation data were available are listed. *DNA methylation data from HumanMethylation27 platform that covers 2 CpGs out of 7 CpGs covered by HumanMethylation450 were used.
| TCGA Cancer Type Name | Abbreviation | Tumor samples | Normal samples |
|---|---|---|---|
| adrenocortical carcinoma | ACC | 79 | 0 |
| cervical squamous cell carcinoma and endocervical adenocarcinoma | CESC | 304 | 3 |
| cholangiocarcinoma | CHOL | 36 | 9 |
| esophageal carcinoma | ESCA | 184 | 9 |
| glioblastoma multiforme | GBM | 51 | 1 |
| kidney chromophobe | KICH | 66 | 0 |
| brain lower grade glioma | LGG | 516 | 0 |
| mesothelioma | MESO | 87 | 0 |
| ovarian serous cystadenocarcinoma | OV* | 295 | 0 |
| pheochromocytoma and paraganglioma | PCPG | 179 | 3 |
| sarcoma | SARC | 259 | 0 |
| stomach adenocarcinoma | STAD | 373 | 0 |
| testicular germ cell tumors | TGCT | 150 | 0 |
| thymoma | THYM | 120 | 2 |
| uterine carcinosarcoma | UCS | 57 | 0 |
| uveal melanoma | UVM | 80 | 0 |
Seven of sixteen analyzed cancer types (CESC, CHOL, ESCA, MESO, SARC, STAD, and UCS) show strong MORT silencing by DNA methylation (Figure 1). The negative correlation rho between MORT expression and DNA methylation in these cancers is below -0.5; the DNA methylation level in some tumor samples of these cancers exceeds 0.5 beta (> 50% DNA methylation), and a large fraction of the tumor samples in these cancer types have very low to no MORT expression level (Figure 1). The correlation of MORT expression and promoter DNA methylation in the remaining nine cancer types is also negative; however, the maximum level of the DNA methylation of MORT promoter in some of these cancers is either very low (UVM), or a very few tumor samples have MORT silenced (ACC, KICH, OV, and THYM), and some of the cancer types (GBM, LGG, PCPG, and TGCT) do not appear to display MORT gene silencing (Figure 1).
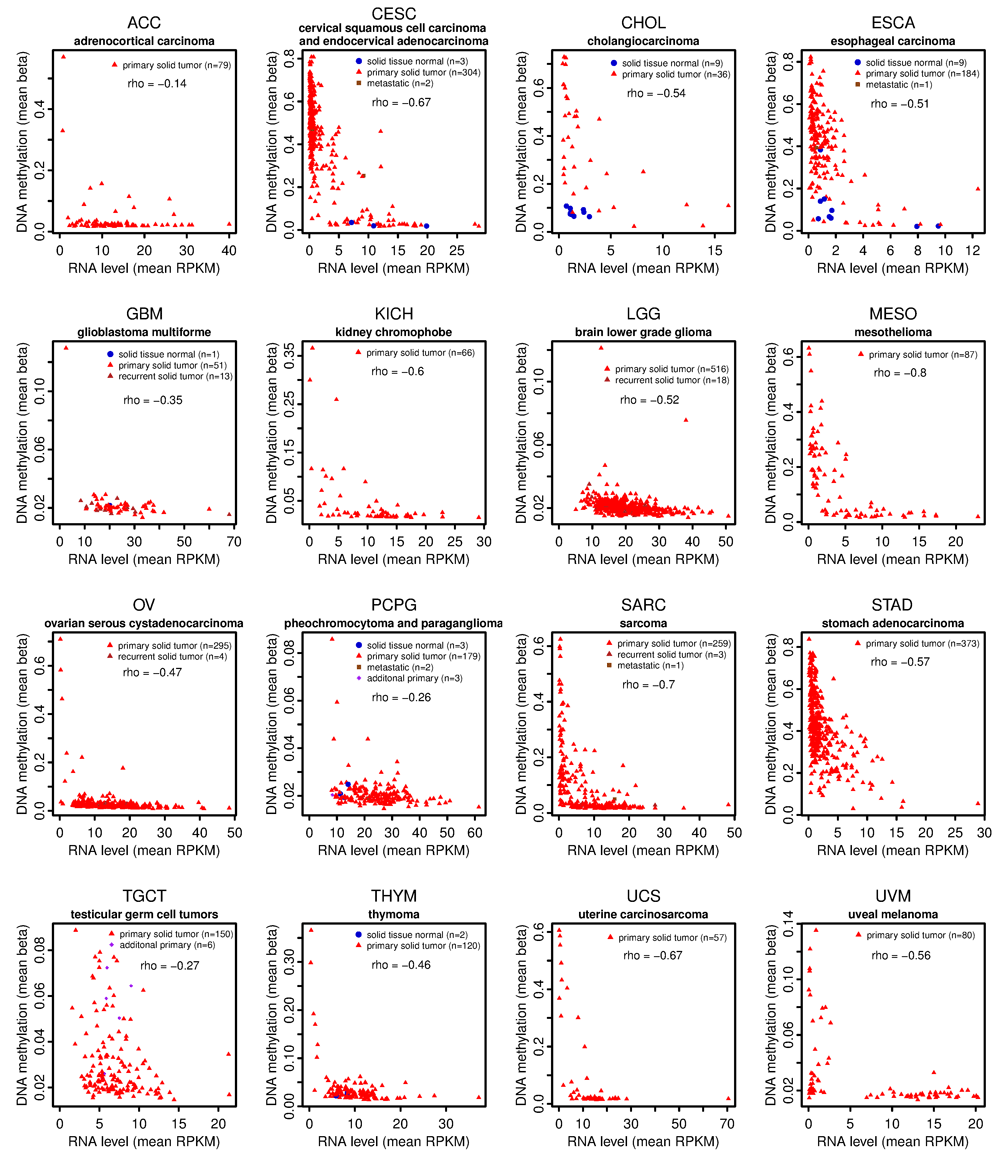
The x-axis shows the MORT expression level according to RNA-seq and y-axis shows the level of MORT promoter DNA methylation according to Illumina HumanMethylation450 microarray. The correlation coefficient rho between the MORT expression and the DNA methylation of MORT promoter for each tumor type is displayed. The OV has a very low number (10) of samples analyzed by the HumanMethylation450 platform, therefore the data from the HumanMethylation27 platform that covers 2 CpGs out of 7 CpGs covered by HumanMethylation450 were used.
The analysis presented shows DNA methylation associated MORT gene silencing in 7 of 16 TCGA cancer types. Compared to the 17 TCGA cancer types presented in our original study,1 most the 16 cancer types presented here lack their respective normal tissues samples and some of them have lower amounts of tumor samples (Table 1). Nevertheless, the distribution of MORT expression and DNA methylation data in tumor samples clearly indicates MORT silencing in multiple cancer types (Figure 1).
Cervical tumors (CESC) have high proportion of MORT silencing (Figure 1); more than 75% of 304 cervical tumor samples have MORT promoter DNA hypermethylated and MORT silenced. Using TCGA data, a recent study found MORT downregulated in cervical cancer11, but surprisingly did not report on or hypothesize potential mechanisms for this transcriptional repression. Here we confirm and extend their initial analysis of MORT silencing in cervical cancer and show further that this silencing is strongly linked to aberrant DNA methylation of the MORT promoter.
Combined together with the findings from our previous report1, Table 2 shows MORT is silenced by DNA methylation in a super majority of TCGA cancer types (22 of 33). MORT loss occurs predominantly due to epigenetic silencing and increased DNA methylation of its promoter in breast cancer9. This could likely be extended to all 22 cancer types with the high fraction of MORT negative samples and the high correlation between MORT RNA level and MORT promoter DNA methylation, where MORT likely plays a tumor suppressive role. The other 11 cancer types, with a little to no MORT silencing, might have tumor suppressive pathway, where MORT is involved, interrupted elsewhere and/or MORT may play some additional vital role in tissues these tumors originate from - e.g. prostate, thyroid, brain, testes, or ovary - since these tissues typically have the highest levels of MORT RNA1.
The cancer types with MORT silencing in a large fraction of tumor samples are indicated. Results from this study are indicated (*), results from our previous report (ref 1) are indicated (**).
| Abbreviation | TCGA cancer type name | MORT silencing |
|---|---|---|
| ACC | adrenocortical carcinoma | No* |
| BLCA | bladder urothelial carcinoma | Yes** |
| BRCA | breast invasive carcinoma | Yes** |
| CESC | cervical squamous cell carcinoma and endocervical adenocarcinoma | Yes* |
| CHOL | cholangiocarcinoma | Yes* |
| COAD | colon adenocarcinoma | Yes** |
| DLBC | lymphoid neoplasm diffuse large b-cell lymphoma | Yes** |
| ESCA | esophageal carcinoma | Yes* |
| GBM | glioblastoma multiforme | No* |
| HNSC | head and neck squamous cell carcinoma | Yes** |
| KICH | kidney chromophobe | No* |
| KIRC | kidney renal clear cell carcinoma | Yes** |
| KIRP | kidney renal papillary cell carcinoma | Yes** |
| LAML | acute myeloid leukemia | Yes** |
| LGG | brain lower grade glioma | No* |
| LIHC | liver hepatocellular carcinoma | Yes** |
| LUAD | lung adenocarcinoma | Yes** |
| LUSC | lung squamous cell carcinoma | Yes** |
| MESO | mesothelioma | Yes* |
| OV | ovarian serous cystadenocarcinoma | No* |
| PAAD | pancreatic adenocarcinoma | Yes** |
| PCPG | pheochromocytoma and paraganglioma | No* |
| PRAD | prostate adenocarcinoma | No** |
| READ | rectum adenocarcinoma | Yes** |
| SARC | sarcoma | Yes* |
| SKCM | skin cutaneous melanoma | Yes** |
| STAD | stomach adenocarcinoma | Yes* |
| TGCT | testicular germ cell tumors | No* |
| THCA | thyroid carcinoma | No** |
| THYM | thymoma | No* |
| UCEC | uterine corpus endometrial carcinoma | Yes** |
| UCS | uterine carcinosarcoma | Yes* |
| UVM | uveal melanoma | No* |
In summary, our findings show that the MORT gene is one of the most common epigenetic aberrations seen in human cancer. Coupled together with MORT silencing occurring early in the temporal arc of human carcinogenesis it strongly supports a tumor suppressive role for MORT.
Illumina HiSeq RNA-seq and HumanMethylation450 DNA methylation data for TCGA cancer types used in the present study can be downloaded from the GDC data portal.
This work was supported by the Maynard Chair in Breast Cancer Epigenomics at the University of Arizona Cancer Center and the Cancer Center Support Grant (P30 CA023074).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The results shown here are based upon data generated by the TCGA Research Network: http://cancergenome.nih.gov/.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: DNA methylation biomarkers, liquid biopsies, digital PCR
Competing Interests: No competing interests were disclosed.
Competing Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |||
|---|---|---|---|
| 1 | 2 | 3 | |
|
Version 1 21 Feb 18 |
read | read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)