Keywords
Diversity, ITS, Llanganates, National Park, Sangay, Xylarial
This article is included in the Ecology and Global Change gateway.
Diversity, ITS, Llanganates, National Park, Sangay, Xylarial
According to the referee's suggestions we have made the following changes in the manuscript:
To read any peer review reports and author responses for this article, follow the "read" links in the Open Peer Review table.
Sangay (SP) and Llanganates (LP) National Parks in Ecuador are considered as high priority conservation units in the Tropical Andes, due to their high biodiversity and high endemism1,2. However, their mycological diversity is still unknown. This study aims to contribute to the conservation of fungi, showing the results of their diversity, based on molecular taxonomy, by analyzing the ITS (internal transcribed spacer) regions. ITS is the accepted as primary fungal barcode marker for fungi3,4. For this, the DNA sequence of specimens of exploratory fungal collections were analyzed within the aforementioned parks. Here we present results exclusively for the Xylariales order, other fungal orders were also collected but are not shown here.
Sample collection was carried out during the months of January and February 2015. The fruiting bodies collected were deposited in the QCAM Fungarium (Catholic University Mycology Collection, Quito). Table 1 displays the collection codes, as stored at the QCAM. The ITS1-5.8SITS2 region was amplified by PCR with primers ITS1F (5’-CTTGGTCATTTAGAGGAAGTAA-3’)5 and ITS4 (5’-TCCTCCGCTTATTGATATGC-3’)6. The amplified fragments were sent for sequencing to Macrogen Inc. (Seoul, South Korea). The forward and reverse sequences obtained for each isolate were assembled in Geneious R8 (Biomatter Ltd. 2005–2012), and submitted to GenBank. Sequence data were analyzed by comparison with sequences available in GenBank. An assignment to the lower taxonomic level was made by direct homology of the consensus sequences with the search results in BLASTn (NCBI) optimized for highly similar sequences (megablast). Alignments that presented 100% coverage and at least 99% identity with a sequence previously reported in GenBank were considered. The results were compared with the previously made morphological identification at the QCAM Fungarium, to check the taxonomic designation.
*SNPs: Single nucleotide polymorphisms found between the sample analyzed and the closest hit on the BLASTn search.
Sequence data were aligned with Geneious R8 and later manually adjusted with Mesquite version 3.047. Public sequences available in GenBank that corresponded to specimens that gave the greatest homology in BLASTn with the sequences of the collected specimens were included. Phylogenetic trees were constructed in Geneious R8 using the PhyML8 plugin for Maximum Likelihood (ML) with a custom substitution model (010230), determined by jModelTest 2.1.4.9,10, according to Corrected Akaike Information Criterion (AICc)11,12. A bootstrap of 1000 replica was used.
All the specimens analyzed were of the genus Xylaria. The eight specimens from Llanganates National Park were identified as X. enterogena, X. fissilis, X. schweinitzii, X. telfairii and three unidentified species. For the two samples from Sangay National Park, one was X. telfairi and the other was an unidentified species. Differences in the number of samples found at each park could be due to the sampling effort, and not necessarily to the richness of the Xylareales in the Parks. The unidentified species were different in each park. The analysis shows that there are no shared species of Xylaria at the two sampled sites (Table 1), this is important for conservation decision making. The phylogenetic relationships recovered from the analysis of the ITS sequences (Figure 1) shows two major groups. The first major group, composed by clades A and B, is well supported, it includes specimens from LP and PS. Clade A includes all X. entogena specimens and Clade B includes all X. telfairii specimens, and Xylarya sp.1 specimens. It is possible that Xylarya sp.1 might belong to the X. telfairii group, but due to the differences among the sequences it is likely a different species. In the second major group, clade C is sister to clades D, E and F. This major group includes specimens from both LP and SP. Clade C includes all X. schweinitzii specimens. Clade D includes Xylaria sp. 2. The closest sequence to Xylaria sp. 2 from SP was a previously reported collection also from Ecuador13 in a cloud forest in the province of Imbabura, that was also identified only at the genus level. Clade E shows Xylaria sp. 3, the closest sequence to this individual belongs to the same previously reported study13, identified only at the genus level. Clade F includes Xylaria fissilis sequences from LP and one from 13. Clade F also includes Xylaria sp. 4, an unidentified specimen. The number of nucleotide differences (SNPs) between the sample and the closest hit on the BLASTn search suggests that these specimens may belong to new species (Table 1). Additional loci and more detailed morphological analyses are needed to determine this. The genus Xylaria is probably the largest in the family Xylariaceae, with 35 estimated genera14, but the real number remains unknown15. Studies related to the biological diversity of this order in the National Parks of Ecuador are scarce, more systematic field studies would surely reveal a greater diversity of families, genera and species within the Xylariales in SP and LP, as well as other regions and protected areas of Ecuador, especially if we take into account the cosmopolitan distribution of Xylaria13. In fact, new fungal species in SP, belonging to the Agaricales, have recently been described16.
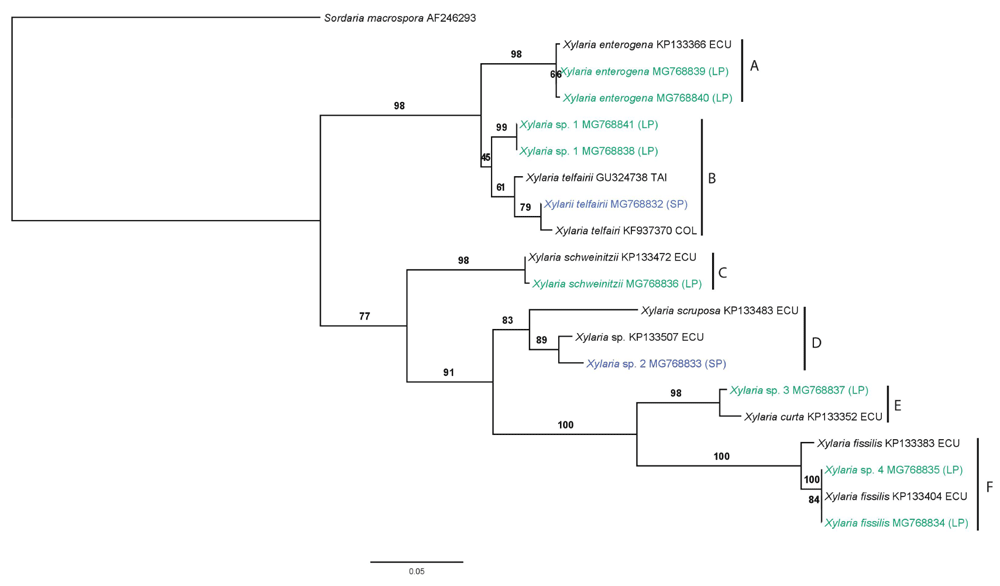
These entire unidentified specimens might represent new species. Additional loci and more detailed morphological analyses are needed to determine this. The genus Xylaria is probably the largest in the family Xylariaceae, with 35 estimated genera13, but the real number remains unknown15. Studies in relation to the biological diversity of this order in the National Parks of Ecuador are scarce, more systematic field studies would surely reveal a greater diversity of families, genera and species within the Xylariales in SP and LP, as well as other regions and protected areas of Ecuador, especially if we take into account the cosmopolitan distribution of Xylaria13. In fact, new fungal species in PS, belonging to the Agaricales, have recently been described16.
The results obtained allow us to establish a baseline for the biological diversity of the Xylariales in SP and LP, an important step to the conservation of fungi. This is the main contribution of this study. We found four species of Xylaria: X. enterogena, X. telfairii, X. schweinitzii, and X. fissilis, and four potential new species based on ITS sequences divergence; the species found in LP are different from those found in SP. However, there is much more to discover. A huge and complex task is pending. To advance our understanding of the Kingdom Fungi we must start by deciphering the diversity of species present in these sites.
The sequencing data are available on the NCBI Genbank webpage: Xylaria enterogena:
https://www.ncbi.nlm.nih.gov/nuccore/MG768840
https://www.ncbi.nlm.nih.gov/nuccore/MG768839
Xylaria fissilis: https://www.ncbi.nlm.nih.gov/nuccore/MG768834
Xylaria schweinitzii: https://www.ncbi.nlm.nih.gov/nuccore/MG768836
Xylaria telfairii: https://www.ncbi.nlm.nih.gov/nuccore/MG768832
Xylaria sp. 1: https://www.ncbi.nlm.nih.gov/nuccore/MG768838
https://www.ncbi.nlm.nih.gov/nuccore/MG768841
Xylaria sp. 2: https://www.ncbi.nlm.nih.gov/nuccore/MG768833
Xylaria sp. 3: https://www.ncbi.nlm.nih.gov/nuccore/MG768837
Xylaria sp. 4: https://www.ncbi.nlm.nih.gov/nuccore/MG768835
The Secretaria de Educación Superior, Ciencia, Tecnología e Innovación del Ecuador (SENESCYT), Arca de Noé Initiative and the QCAM Fungarium from "Pontificia Universidad Católica del Ecuador" supported the project.
We are grateful to Marcel A. Caminer, Charles W. Barnes and Cristina E. Toapanta for their contribution to the development of the present research. This research has been carried out according to the MAE-DNB-ARRGG-CM-2014-002 "Contrato Marco de Acceso a Recursos Genéticos".
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Competing Interests: No competing interests were disclosed.
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Phylogenetics
Competing Interests: No competing interests were disclosed.
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Not applicable
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Partly
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Phylogenetics
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |||
|---|---|---|---|
| 1 | 2 | 3 | |
|
Version 2 (revision) 17 May 18 |
read | read | read |
|
Version 1 23 Feb 18 |
read | read | |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)