Keywords
education, biochemistry lab, protein-ligand interactions, mass spectrometry, ribonuclease A
education, biochemistry lab, protein-ligand interactions, mass spectrometry, ribonuclease A
I am very grateful to the reviewers for their comments and suggestions. In this new version of the manuscript, I have expanded the Conclusions section as requested by Dr. Allen. Two sentences were added that refer to centrifugal desalting as a method that could have reduced phosphate adduct formation and would be useful in training students. Five more sentences were added to address how non-ideal ionization conditions and non-specific binding could have affected the measurements. The expansion necessitated inclusion of a new reference (Benkestock et al., 2004) suggested by Dr. Allen.
To read any peer review reports and author responses for this article, follow the "read" links in the Open Peer Review table.
dC5 deoxyoligonucleotide with the sequence: CCCCC
dC2AC2 deoxyoligonucleotide with the sequence: CCACC
RNase A bovine pancreatic ribonuclease A
ESI-IT-MS electrospray ionization ion-trap mass spectrometry
nESI-Q-TOF-MS nanoelectrospray ionization quadrupole time-of-flight mass spectrometry
RNase A+dC5 ligand-bound form of RNase A (with one dC5 ligand)
RNase A+dC2AC2 ligand-bound form of RNase A (with one dC2AC2 ligand)
RSD relative standard deviation
Bovine pancreatic ribonuclease A (RNase A) is an endoribonuclease (EC 3.1.27.5) that hydrolyzes RNA. It is a small single chain polypeptide (124 amino acids) containing four disulfide bridges and is known for its significant stability1. RNase A has been called “the most studied enzyme of the 20th century” and it has seen wide use as a model protein in biochemical and biophysical experiments1. Undergraduate life-science majors often learn of RNase A as part of a biochemistry course in the context of the Nobel Prize winning protein folding experiments performed by Christian Anfinsen2. Students may also be familiar with the need to inhibit ribonucleases when working with RNA in the lab, often accomplished with diethyl pyrocarbonate, or will have learned about the role of ribonucleases in microRNA biology3. Still others may recognize RNase A as an example of an enzyme that employs general acid-base catalysis as part of its chemical mechanism4. Thus, RNase A is an excellent model for undergraduate lab experiments, not only because it has been extensively studied, but also because its use presents an opportunity to reemphasize important concepts in biochemistry and biology.
The application of mass spectrometry to the analysis of biomolecules has made an enormous impact in the life sciences. Protein identification, the characterization of protein modifications, and the quantification of biomolecules using mass spectrometry are commonplace. Of these, protein identification is the most established in an undergraduate teaching lab5–10. Numerous other biological applications of mass spectrometry have existed for many years, but some of these are arguably, less broadly appreciated, and this is especially true for undergraduates. Native mass spectrometry is an approach based on electrospray ionization, where biomolecules are sprayed from a non-denaturing solvent11. Under such conditions, protein-ligand complexes can be maintained and a dissociation constant (Kd) can be determined via a titration experiment12–14.
Previously, nanoelectrospray ionization quadrupole time-of-flight mass spectrometry (nESI-Q-TOF-MS) was used to investigate ligand binding to RNase A12,15,16. These studies used nESI ionization for its superior sensitivity and relied on the TOF mass analyzer for its high mass range12,15,16. In Zhang et al., free RNase A and the ligand-bound forms of RNase A populated three charge states (+8, +7, and +6) at pH 6.6, with most of the signal (~90%) coming from the +7 charge state, which exceeded m/z 2000 in the ligand-bound forms12. Similarity, in Sundqvist et al., focus was placed on the +7 charge state of free RNase A and its ligand-bound forms15. In contrast, Yin et al. reported the most abundant charge state of free and ligand-bound forms of RNase A to be +8 at pH 6.616. Unfortunately, California State University-Chico does not own a nESI-Q-TOF-MS as employed by each of these research groups. Instead, we have an electrospray ionization ion-trap mass spectrometer (ESI-IT-MS), which by comparison to nESI-Q-TOF-MS, offers a lower sensitivity and mass range (50–2000 m/z). Consequently, at the outset of this preliminary investigation, it was recognized that observation of the +7 and +6 charge states of ligand-bound RNase A would not be possible with our instrument.
This work was an attempt to develop a biochemistry lab experience that would introduce undergraduate life-science majors to the use of mass spectrometry for the analysis of protein-ligand interactions. Two deoxyoligonucleotides, CCCCC (dC5) and CCACC (dC2AC2), were investigated for their ability to bind RNase A. Titration experiments were performed using a fixed RNase A concentration and variable deoxyoligonucleotide concentrations. Samples at equilibrium were infused directly into our ESI-IT-MS under native conditions. The relative simplicity of the sample preparation and instrument operation (by direct infusion) were viewed as desirable features for an undergraduate teaching lab. Data analysis was also straightforward. Herein is described the results of this preliminary investigation. This work differentiates itself from the abovementioned RNase A ligand binding studies (using mass spectrometry) by the experimental conditions employed, which includes the identity of the investigated ligands and the type of mass spectrometer used12,15,16.
A stock solution of bovine pancreatic ribonuclease A (#R6513, Sigma-Aldrich, St Louis, MO, USA) was prepared at 5.60 mg/mL in LC-MS grade water (Thermo-Fisher Scientific, Waltham, MA, USA). Ammonium acetate (NH4OAc) was LC-MS grade (#73594, Sigma-Aldrich). HPLC-purified deoxyoligonucleotides with the sequence “CCCCC” (dC5) and “CCACC” (dC2AC2) were obtained from ThermoFisher and the stock solutions (200 μM) were prepared in LC-MS grade water. Samples were prepared in 1.5 mL microcentrifuge tubes as indicated in Table 1. Six replicates were prepared and analyzed for “Sample 1” whereas “Samples 2–5” were prepared and analyzed in triplicate. Each sample was mixed by micropipetting, and incubated at room temperature for ten minutes, prior to analysis.
| Component | Sample # | ||||
|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | |
| RNase A (5.60 mg/mL)1 (μL) | 10 | 10 | 10 | 10 | 10 |
| LC-MS grade H2O (μL) | 40 | 37.5 | 35 | 30 | 20 |
| 20 mM NH4OAc, pH 6.00 (μL) | 50 | 50 | 50 | 50 | 50 |
| 200 µM deoxyoligonucleotide2 (μL) | 0 | 2.5 | 5 | 10 | 20 |
| Total Volume (μL) | 100 | 100 | 100 | 100 | 100 |
| Overall [deoxyoligonucleotide2] (μM) | 0 | 5 | 10 | 20 | 40 |
| Overall [RNase A] (μM) | 40.9 | 40.9 | 40.9 | 40.9 | 40.9 |
1409 μM RNase A; calculated with the MWav (13,690.3) for PDB ID:1RTA (Ref. 17).
2Either dC5 or dC2AC2.
Samples were analyzed with a Thermo LCQ Advantage ion-trap mass spectrometer equipped with an electrospray ionization source. The instrument was operated in positive ion mode using a 4.5 kV spray voltage, 60°C capillary temperature, 200 ms inject time, 10 microscans, and nitrogen sheath and aux gas settings of 30 and 15, respectively. The instrument was tuned on the +8 charge state of free RNase A at m/z 1723.7 (Table 2). Each sample was subjected to direct-infusion at 2.5 µL/min using the LCQ syringe pump and full-scan mass spectra (m/z 1500-1950) were collected for two minutes. The upper m/z range was capped at 1950 to exclude the +7 charge state of free RNase A, which in its various adduct forms, began at m/z 1955.5 (Table 2). The rationale was that the +7 charge state of the ligand-bound forms of RNase A were above m/z 2000, which made +7 data incomplete and unusable (Table 3).
The +8 charge state used in this work is highlighted.
The +8 charge state used in this work is highlighted.
To facilitate determination of total ion abundance, tables of predicted m/z values for free RNase A (Table 2) and the ligand-bound forms of RNase A (RNase A+dC5 and RNase A+dC2AC2) (Table 3) were constructed. A series of 98 Da adducts were included in Table 2 and Table 3 due to their presence in the mass spectra of this work, and that of earlier studies12,15. These adducts have been suggested to be either H2SO4 or H3PO418. Other RNase A studies have assigned these adducts as phosphate, and so each 98 Da adduct (X) in this work was designated as “Pi” (Table 2 and Table 3)12,15. Although mass spectra showed that free RNase A had up to 8 Pi adducts (Figure 1A and 1F), only the 0-5 Pi adduct forms of free RNase A and its ligand bound forms were used. This restraint was necessitated by the predicted m/z overlap of the ligand-bound forms of RNase A (with Pi adducts >5) with the m/z of free RNase at the +7 charge state. The “Qual Browser” feature of Xcalibur 1.4 SR1 software (Thermo) was used for analysis of each *.raw file. For each sample, mass spectra comprising the two-minute data collection were averaged. The “spectrum list view” was used to obtain intensity data for all of the ions in the ranges comprising the +8 charge state (with 0-5 Pi adducts) for free RNase A (m/z 1710.7-1772.9), RNase A+dC5 (m/z 1883.7-1945.9), and RNase A+dC2AC2 (m/z 1886.7-1948.9). The intensity data for all ions in each m/z range were added to give the “total ion abundance” of the free (Ab(P)) and ligand-bound forms (Ab(PL)) of RNase A. The total ion abundance for the ligand-bound forms (RNase A+dC5 and RNase A+dC2AC2) were plotted as a function of [deoxyoligonucleotide] using GraphPad Prism 7.
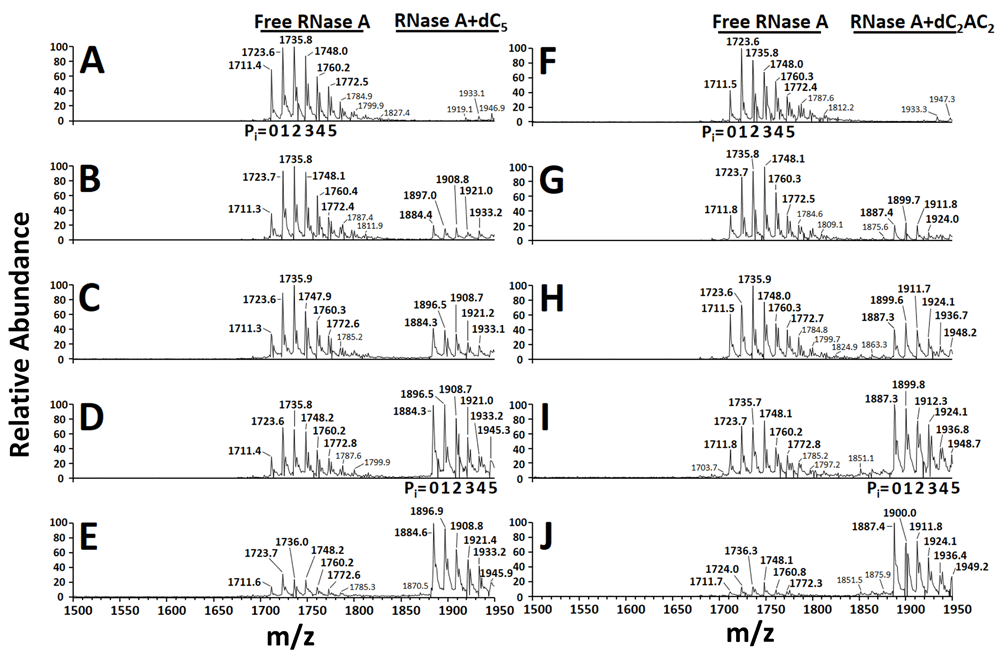
The +8 charge state is shown. (A & F) no added deoxyoligonucleotide, (B) 5 μM dC5, (C) 10 μM dC5, (D) 20 μM dC5, (E) 40 μM dC5, (G) 5 μM dC2AC2, (H) 10 μM dC2AC2, (I) 20 μM dC2AC2, and (J) 40 μM dC2AC2. The number of phosphate adducts (Pi= 0-5) are indicated in four representative mass spectra (A, D, F, and I).
The total ion abundance ratio was determined at each [deoxyoligonucleotide] using the method described by Kitova et al.13, where for a 1:1 protein-ligand complex, the total ion abundance ratio (R) is calculated using the total abundance of all ligand-bound ions (Ab(PL)) and the total abundance of all free protein ions (Ab(P)) as shown in Equation 1:
R= Ab(PL)/Ab(P) = [PL]eq/[P]eq [1]
The total ion abundance ratio (R) is used with the initial ligand concentration ([L]0) and initial protein concentration ([P]0) to calculate the association constant (Ka) using Equation 213:
Ka=R/([L]0 − ((R/(1+R))[P]0)) [2]
The Kd can then be calculated as the reciprocal of the Ka value.
Table 1 indicates that samples contained an overall [RNase A] of 40.9 μM. Relatively low signal intensities observed for the +8 charge state of free and ligand-bound forms of RNase A necessitated this concentration, which was higher than the 5–20 μM RNase A used by others in nESI-Q-TOF-MS experiments12,15,16. Table 2 and Table 3 present predicted m/z values for free RNase A and the ligand-bound forms of RNase A (RNase A+dC5 and RNase A+dC2AC2) with multiple Pi adducts, which correlated well with observed m/z values (Figure 1). Upon increasing the concentration of dC5, the total ion abundance of free RNase A was found to decrease in intensity while the total ion abundance of RNase+dC5 was found to increase in intensity, which suggested 1:1 stoichiometry for the dC5:RNase A interaction (Figure 1A–E). Similar results were seen for the titration using dC2AC2 (Figure 1F–J). Table 4 presents total ion abundance data for free RNase A in samples that contained no added deoxyoligonucleotide. Total ion abundance data for free RNase A across six replicates gave a RSD of 16.4% (Table 4). Table 5 contains total ion abundance data for free RNase A and the ligand-bound forms of RNase A in samples that contained various concentrations of dC5 or dC2AC2. Total ion abundance data across three replicates at each [deoxyoligonucleotide] exhibited RSD values of approximately 20% or less (Table 5). A plot of the total ion abundance for free RNase A, RNase A+dC5, and RNase A+dC2AC2 as a function of [deoxyoligonucleotide] is shown in Figure 2. The total ion abundance for RNase A+dC5 and RNase A+dC2AC2 increased until 20 μM deoxyoligonucleotide, but decreased at 40 μM (Figure 2). Table 6 presents the calculated total ion abundance ratio (R) and dissociation constant (Kd) at each [deoxyoligonucleotide]. Samples containing <40 μM deoxyoligonucleotide unexpectedly produced negative Kd values (Table 6). By contrast, Table 6 shows that samples containing 40 μM deoxyoligonucleotide produced consistent positive values where the average Kd for dC5 was 2.2 ± 0.1 μM and the average Kd for dC2AC2 was 1.0 ± 0.1 μM.
Data is for the +8 charge state.
| Replicate | Free RNase A |
|---|---|
| 1 | 71,438,882 |
| 2 | 80,188,529 |
| 3 | 70,622,004 |
| 4 | 94,929,471 |
| 5 | 61,169,836 |
| 6 | 65,198,871 |
| Average | 73,924,599 |
| SD | 12,135,483 |
| %RSD | 16.4 |
Data is for the +8 charge state.
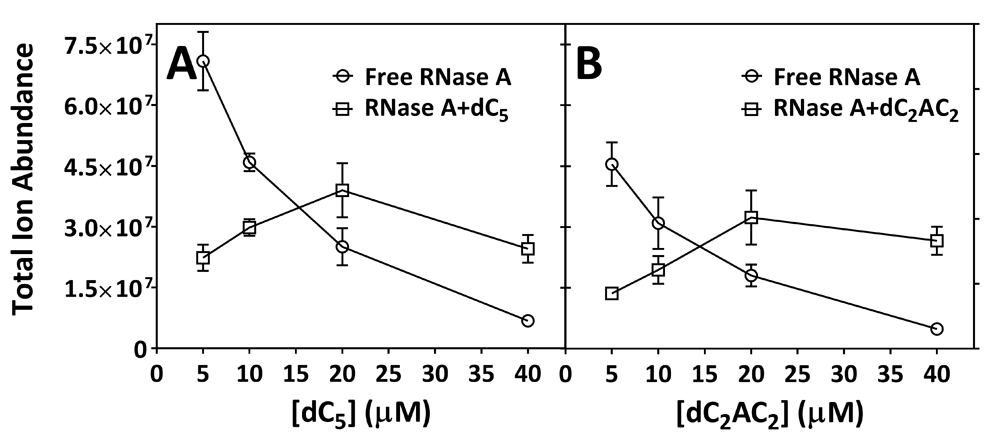
(A) [dC5] and (B) [dC2AC2]. The data is from Table 5, where points represent the average (n=3) ± standard deviation.
Data used for calculations was from Table 5.
This preliminary work demonstrates the potential and pitfalls of a LCQ ESI-IT-MS instrument to investigate protein-ligand interactions in an undergraduate teaching lab. Even though dC5 and dC2AC2 binding to RNase A are clearly illustrated in Figure 1, the presence of the Pi adducts complicated the mass spectra and broadened the signals for free RNase A and the ligand-bound forms of RNase A. In-source collision-induced dissociation was explored to reduce Pi adduct formation, but it appeared to disrupt the RNase A+dC5 and RNase A+dC2AC2 complexes, and so this approach was abandoned (data not shown). Although it was not attempted, centrifugal desalting of the RNase A stock solution might have eliminated Pi adducts and improved the quality of the mass spectra in Figure 1. As an added benefit, in the context of an undergraduate lab, desalting would also introduce students to a common sample preparation technique. It is unclear why the decrease in the total ion abundance for the ligand-bound forms of RNase A was observed at higher deoxyoligonucleotide concentrations (Figure 2). Previously, the ion intensity ratio of free RNase A to the RNase A+cytidine 2′-monophosphate (2′-CMP) complex was observed to vary with charge state as follows: +8 (0.65), +7 (0.73), +6 (1.1)12. This led Zhang et al. to suggest that either the binding of ligand, or the presence of ligand in the analyzed RNase A samples, created a change of the charge state distribution for the protein-ligand complex12. In the present work, the binding of deoxyoligonucleotide, or the presence of deoxyoligonucleotide in samples, could have shifted some of the total ion abundance of free and/or ligand-bound RNase A from the +8 charge state to lower charge states, which were beyond the mass range of our ion-trap mass analyzer. This highlights an inherent limitation of this work, which was the inability to gather data for all free and ligand-bound RNase A charge states. Kitova et al. stated the importance of including all ligand-bound and free protein ions in the calculation of R, and emphasized that the “sometimes-used practice” of employing a particular charge state to determine Ka should be avoided13. Thus, the lack of data for the +7 and +6 charge states of RNase A hindered accurate collection of total ion abundance data, which may have affected calculations of R and led to the negative Kd values at low ligand concentrations (Table 6). Other factors to consider, that could have affected measurements, include non-ideal ionization conditions and non-specific ligand binding. Benkestock et al. showed that instrument-derived parameters (e.g. capillary-to-cone distances) could affect the protein-ligand complex to free protein ratio19. They also demonstrated that compared to pneumatically assisted ESI, which was used in this work, nESI better reflects the equilibrium between free protein and protein–ligand complexes in solution. Furthermore, Kitova et al. noted that changes in the magnitude of Ka, with changes in ligand concentration, might indicate nonspecific ligand binding13. As seen in Table 6, Kd values varied with the deoxyoligonucleotide concentration. Therefore, it is reasonable to suspect that non-specific binding may have contributed to the decreased total ion abundance of the ligand-bound forms of RNase A at higher ligand concentrations (Figure 2). Notwithstanding these possibilities, the positive Kd values in Table 6 are of similar magnitude to those determined by Zhang et al. for 2′-CMP and CTP, via a nESI-Q-TOF-MS titration experiment, which were 1.7 ± 0.3 μM and 0.8 ± 0.2 μM, respectively12. They are also in the neighborhood of in solution Kd measurements (3-24 μM) observed for the binding of short fluorescein-labeled deoxyoligonucleotides to RNase A20. In conclusion, while RNase A is an excellent model for many experiments, instructors wishing to use a LCQ ESI-IT-MS instrument to investigate protein-ligand interactions are encouraged to consider other protein-ligand systems that would enable all charges states (of the free and ligand-bound protein) to be observed.
Dataset 1. LCQ *.raw data files for all samples. 10.5256/f1000research.14268.d19837321
Data files 1–6 are for samples that contained free RNase A (6 replicates), Data files 7–18 are for samples that contained RNase A and dC5 (3 replicates per [dC5]), Data files 19–30 are for samples that contained RNase A and dC2AC2 (3 replicates per [dC2AC2]).
This work was supported by the College of Natural Sciences and the Department of Chemistry and Biochemistry at California State University- Chico.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
I thank Professor Daniel Edwards at California State University-Chico for helpful discussions and review of the manuscript.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Native Mass Spectrometry
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Biochemistry, Structural Biology
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Partly
References
1. Going CC, Xia Z, Williams ER: New supercharging reagents produce highly charged protein ions in native mass spectrometry.Analyst. 2015; 140 (21): 7184-94 PubMed Abstract | Publisher Full TextCompeting Interests: No competing interests were disclosed.
Reviewer Expertise: Native Mass Spectrometry
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 2 (revision) 26 Apr 18 |
read | |
|
Version 1 20 Mar 18 |
read | read |
Click here to access the data.
Spreadsheet data files may not format correctly if your computer is using different default delimiters (symbols used to separate values into separate cells) - a spreadsheet created in one region is sometimes misinterpreted by computers in other regions. You can change the regional settings on your computer so that the spreadsheet can be interpreted correctly.
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)