Keywords
Hookworm, Genotyping, Molecular sequencing, Necator americanus, Sudan
This article is included in the Pathogens gateway.
Hookworm, Genotyping, Molecular sequencing, Necator americanus, Sudan
Ancylostomatidae and strongyl nematodes were known to pose a burden among a variety of mammalian hosts, including humans1. Ancylostoma duodenale and Necator americanus are the species recorded as being most responsible for human infection. They are found in mostly in warm and tropical areas2, and infect around 1.3 billion people worldwide3. A. duodenale are located in Central, Eastern and Northern Africa, India, Australia and Europe4. Further, N. americanus is present in Sub-Saharan Africa, Eastern Asia and Southeast Asia4. A zoonotic species in Asia (A. ceylanicum), also causes human infection; however, this is of limited significance and its exact geological appropriation has not been depicted. The most widespread of all hookworms are A. caninum, a parasite that infects dogs, and has lately been affirmed to survive in the human gut (but without developing sexually)5. Laboratory diagnosis of hookworm infection, routinely based on the presence of eggs in faeces by direct microscopy and/or concentration techniques6. Sometimes after gathering, through faeces culture tests (after 24 hours), hookworm ova may have hatched and rhabditiform larvae might be visible; due to morphological similarity between the two, these larvae must be differentiated from Strongyloides larvae. The severity of disease can depend on the number of ova that are counted in faeces. Furthermore, in some cases, adult hookworms may be found6. Use of molecular techniques (such as PCR) for parasitic detection and identification is more accurate and effective than conventional methods, and requires DNA of the hookworms for detection. For example, identifying both types of internal transcript spacer (ITS1 and ITS2, individually) of ribosomal DNA (rDNA) are proven genetic markers of parasitic nematodes, including A. duodenale and N. americanus.
The aim of this study was to estimate the prevalence and molecular characterization of hookworms isolated from the stool of food handlers attending Public Health Laboratories in Khartoum state, Sudan, for annual check-ups.
A total of 350 stool samples were collected. A previously described formula was used to determine sample size7.
Food handlers working in food facilities in Khartoum State, annually medically checked in public health laboratories and willing to participate were included in this study irrespective of their age, gender and nationality. The public health laboratories were located in Khartoum State, Omdurman locality, Khartoum North locality and Khartoum locality, with sample collection conducted between October 2016 and April 2017. The age of participant ranged from 16 to 68 years, with an average age of 32 years; 46% of the participants were less than 29 years old compared with 54% of being 29 or above. The majority of participants were males (83.19%), with 16.81% being female. Distribution of samples according to the residence data showed that 101 participants were from Khartoum north (28.9%), 160 participants were from Omdurman (45.7%) and 89 participants were from Khartoum (25.4%).
In the first stage, the collected specimens were examined using a microscope (Olympus CX22, Japan), the formol-ether concentration technique and Baermann’s technique, as described previously8. Positive detected samples (those that included hookworm ova/larvae) were examined using PCR and DNA sequencing techniques for genotyping. Fresh stool specimens were collected from public health labs in Khartoum State.
Samples that were found to be positive for hookworms eggs by direct examination, or for larvae by Baermann’s technique, were selected for PCR testing (five samples). The stool samples that were negative for parasites by direct smear or the formol-ether concentration technique on three consecutive stool samples were used as negative controls. For molecular examinations, all stool samples were preserved in 70% ethanol at −20°C. The third-stage larvae that were recovered by Baermann’s technique (Filariform) were collected and preserved. The extracted DNA from filariform larvae were used as control DNA during the molecular assay using Biotechnology G-Spinтм Total DNA Extraction Kit (iNtRON Biotechnology, Inc.), according to the manufacturer’s protocol.
DNA was extracted from stool as per manufacturer's instruction used iNtRON Biotechnology G-Spinтм Total DNA Extraction Kit (iNtRON Biotechnology, Inc.) according to the manufacturer’s instructions.
One primer pair was used: RTHW1F (forward): 5'-GAT GAG CAT TGC WTG AAT GCC G-3') and RTHW1R (reverse): 5'-GCA AGT RCC GTT CGA CAA ACA G-3'9.
The partial ITS1, full-length 5.8S gene, and partial ITS2 ribosomal DNA regions were amplified from larvae and ova using PCR. Amplicon sizes were approximately 485 bp (if it typical to N. americanus) or 380 bp (if it typical to Ancylostoma spp.). The procedure for single-round PCR amplification was performed according to Maxime PCR premix kit (iNtRON Biotechnology, Inc.) REF technique. Briefly 5 µl of DNA extract was added to PCR premix (Maxime PCR premix kit i-Taq), containing i-TagTM DNA polymerase, dNTP mixture and reaction buffer. Next, 2 µl primer (forward and reverse) was added alongside 13 µl of nuclease-free water. The reaction mixture was initially denatured at 94°C for 5 min, followed by 30 cycles of denaturation at 94°C for 30 secs, annealing at 65°C for 30 secs and extension at 72°C for 30 sec. This was followed by a final extension step for 10 min at 72°C in a thermal cycler (SensoQuest GmbH, Germany).
Approximately 5µl of PCR product was elecrophoresed on a 1.5% agarose gel (containing 1.5 µg/100 ml ethidium bromide) in Tris-borate-EDTA buffer, along with the tracking dye bromophenol blue, initially at 120 V and 35 A for 60 min. Thereafter, bands were visualized under UV light and an amplicon of 485 bp was considered positive for hookworm DNA.
DNA sequencing was carried out to confirm identification of the pathogen. Owing to the limited amount of DNA generated from one sample, only four samples of PCR products (485 bp) were sequenced using Sanger sequencing (Macrogen, Inc., Korea). The DNA fragment was 485 bp (if from N. americanus) or 380 bp (if it typical of Ancylostoma spp) from an internal sequence of the amplicon of single-round PCR were obtained using the specific primers. Two sequence fragments were generated for five samples, which were edited manually to correct possible base errors using BIOEDIT 7.09. They were then subsequently joined to reconstruct a fragment of 485 bp or 380 bp spanning genes for hookworm spp.
DNA sequences were compared with the NCBI database to check DNA sequencing quality and specificity using the nucleotide BLAST server. Sequences were submitted in sequence alignment form to Clustal W (online tool) for multiple sequence alignment.
Firstly, before uploading the sequences to NCBI, we proofread the nucleotide chromatogram using Finch TV software version 1.4.0 to ensure that all ambiguous sites were correctly called and to determine the overall quality. Next, nucleotides sequences were searched for sequence similarity using nucleotide BLAST10 Highly similar sequences were retrieved from NCBI and subjected to multiple sequence alignment using BIOEDIT software11.
In this study, the stool samples of 350 participants were investigated for hookworms; five samples were found to be positive (1.43%) using the formol-ether concentration technique. One sample was found to be positive using Baermann’s technique, which was used as the positive control. Five samples were found to positive by PCR, as shown in Figure 1.
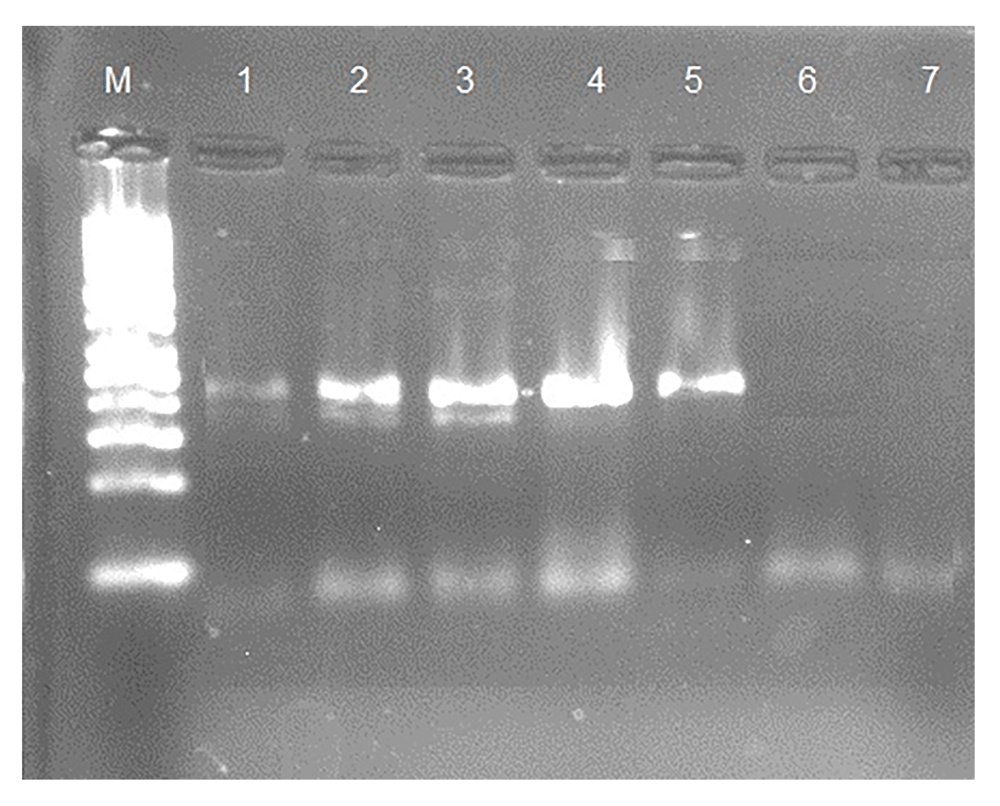
Lane M, 100 bp marker; lane 1, postive control 485 bp; lanes 2–5, postive samples; lane 6, negtive control; lane 7, negative sample.
Four hookworm samples (91, 92, 294 and 319) were sequenced by Macrogen, Inc., Korea and the sequences uploaded to NCBI Genbank (accession numbers: MH035824 (sample 91), MH035825 (sample 92), MH035826 (sample 294) and MH035827 (sample 319). Using nucleotide BLAST, the sequence of samples 91, 92, 294 and 319 were queried for alignment. Samples 91 and 92 showed 100% similarity with N. americanus isolated genes for 18S rRNA, ITS1, 5.8S rRNA, ITS2 and 28S rRNA (Figure 2). Sample 294 showed 98% similarity to N. americanus isolated genes for 18S rRNA, ITS1, 5.8S rRNA, ITS2 and 28S rRNA (Figure 3). Sample 319 showed 97% similarity with N. americanus isolated genes for 18S rRNA, ITS1, 5.8S rRNA, ITS2 and 28S rRNA, (Figure 4).
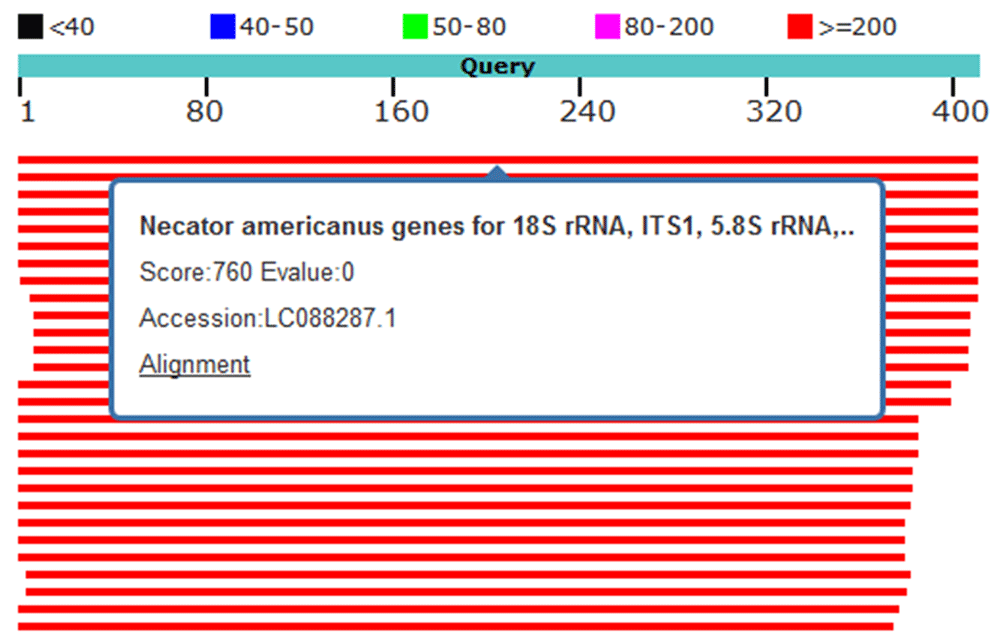
Red lines indicate high identity; sample 91 showed 100% identity to Necator americanus genes for 18S rRNA, ITS1, 5.8S rRNA and ITS2.
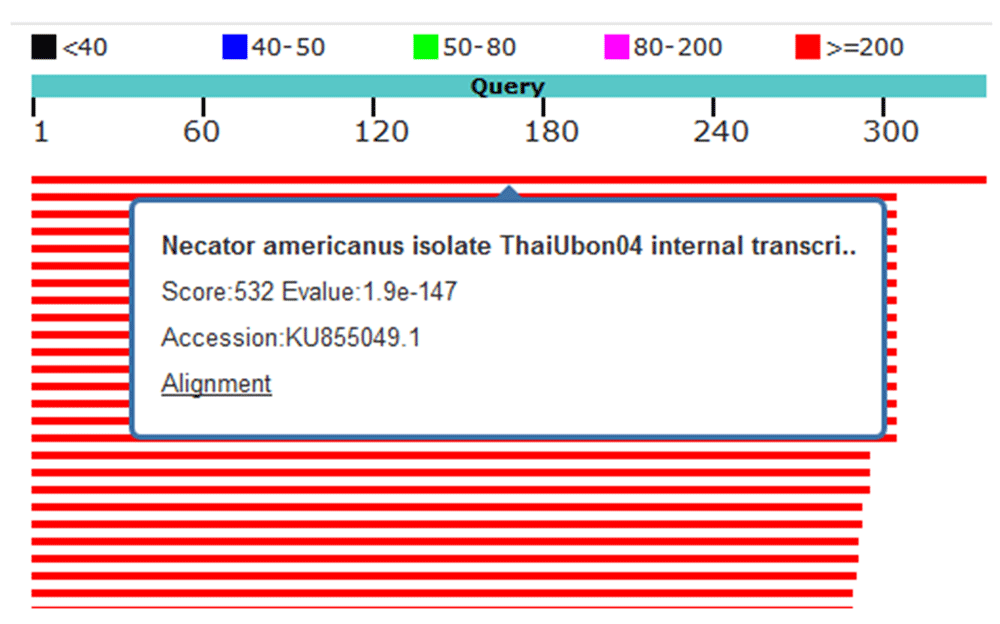
Red lines indicate high identity. Sample 294 showed 98% identity to Necator americanus genes for 18S rRNA, ITS1,5.8S rRNA and ITS2.
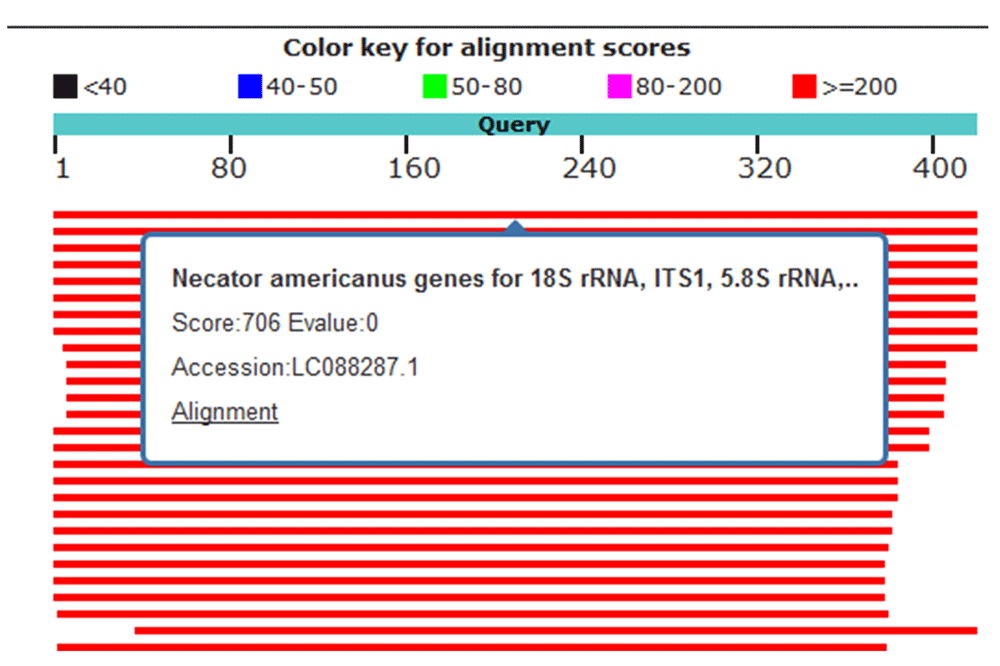
Red lines indicate high identity. Sample 319 showed 97% identity to Necator americanus genes for18S rRNA, ITS1,5.8S rRNA and ITS2.
The present study indicates that the prevalence of hookworms in food-handlers who attended for annual check-ups in Khartoum State, Sudan was 1.43%. To the best of our knowledge, this is the first study to identify hookworm infection in Sudan using molecular techniques; it can therefore serve as a baseline for studies of hookworms in Sudan. Molecular techniques are more advantageous for hookworm identification as they are rapid and more sensitive. DNA was extracted from larva and ova using the iNtRON Biotechnology G-Spinтм Total DNA Extraction Kit. Partial ITS1, full-length 5.8S and partial ITS2 rDNA regions were amplified and then sequenced. Molecular analysis was used to confirm human infections with one species of human hookworms, namely, N. americanus. The nucleotide sequences were analyzed by nucleotide BLAST searching and DNA was aligned using Clustal W13. Amplicon sizes were approximately 485 bp (typical of N. americanus). Sequences showed extremely high similarities (97–100%) with hookworm sequences in the GenBank database. The 4 samples studied were N. americanus. They were similar to N. americanus LC036565.1 (Japan), KM891738.1 (China) and LC036563.1 (Southern Vietnam). All sequences obtained in this study were deposited in the GenBank database. The results also indicate that PCR may be considered as a sensitive method for confirming a diagnosis of hookworm infection, and can aid the clinician in initiating prompt and appropriate antiphrastic therapy.
The differences in hookworm species populations causing human disease in different areas may possibly be related to parasite manner, ethnicity, atmosphere, temperature, and ecological factors2,14. The finding of N. americanus in Sudan goes well with other finding reported from Peninsular Malaysia15 and Thailand9, where N. americanus was more common than A. ceylanicum. Their findings also supported this study that A. duodenale infection was not found.
PCR is a valuable tool for the laboratory diagnosis of parasites. It was found to be effective, sensitive and confirmatory for the diagnosis of hookworm infection and can aid the clinician in initiating prompt and appropriate antiparasite therapy. We confirmed that the major hookworm species infecting humans in Sudan is N. americanus.
Dataset 1. Complete positive/negative results for each technique used to identify parasites in every stool sample. DOI: 10.5256/f1000research.14683.d20417612.
Sequence of sample 91, Accession number MH035824: http://identifiers.org/ncbigi/GI:1356678983.
Sequence of sample 92, Accession number MH035825: http://identifiers.org/ncbigi/GI:1356678984.
Sequence of sample 294, Accession number MH035826: http://identifiers.org/ncbigi/GI:1356678985.
Sequence of sample 294, Accession number MH035827: http://identifiers.org/ncbigi/GI:1356678986.
The authors extend their heartfelt thanks to the participants and staff for their support.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Partly
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Disease ecology, wildlife pathology, theoretical disease ecology, vector borne and zoonotic diseases
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Partly
If applicable, is the statistical analysis and its interpretation appropriate?
Not applicable
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 24 May 18 |
read | read |
Click here to access the data.
Spreadsheet data files may not format correctly if your computer is using different default delimiters (symbols used to separate values into separate cells) - a spreadsheet created in one region is sometimes misinterpreted by computers in other regions. You can change the regional settings on your computer so that the spreadsheet can be interpreted correctly.
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)