Keywords
Crustacean, homeobox, TALE, comparative genomics, arthropod, homeodomain
Crustacean, homeobox, TALE, comparative genomics, arthropod, homeodomain
As one of the fastest growing industries, the seafood trade is dominated by fishing and farming of crustaceans, with annual sales exceeding $40 billion (Stentiford et al., 2012). Crustacean aquaculture is multi-faceted, not only contributing to the ever-increasing demands by international markets, but is also directly linked to the socio-economic aspects of many developing nations through the creation of jobs and infrastructure. Aquaculture practices have intensified in recent years to cope with the demand. Yet, many are not sustainable since the increased densities of farmed shrimps often serve as hotbeds for pathogens if left unabated, causing infectious diseases and the devastation of cultures resulting in massive financial losses. As a result, regulations associated with aquaculture diseases are being enforced with emphasis placed on preventative measures, e.g. enhancement of broodstock and research aiming to further our understanding on crustacean development and ways to utilize the innate ability of crustaceans to combat pathogens (Lai & Aboobaker, 2017; Stentiford et al., 2012).
Several conserved molecular genetic circuitries are well-known for regulating many aspects of development and innate immune homeostasis. One prominent example would be homeobox genes, a family of transcription factors defined by the presence of a homeodomain (Holland, 2013). As one of the most important master controls in development, some headway has already been made in understanding the involvement of homeobox genes in innate immunity; Caudal in Drosophila melanogaster is implicated in commensal-gut mutualism (Ryu et al., 2004; Ryu et al., 2008). Given their importance, major efforts have thus far focused on characterization of homeobox genes in well-known model organisms such as humans (Garcia-Fernàndez, 2005; Holland et al., 2007), Caenorhabditis elegans (Bürglin, 1997), D. melanogaster (Mukherjee & Bürglin, 2007), planarians (Currie et al., 2016; Felix & Aboobaker, 2010; Garcia-Fernandez et al., 1991), amphioxus (Luke et al., 2003), teleost fish (Mulley et al., 2006) and many more. Although homeobox orthologs have been previously studied in the crustacean Parhyale hawaiensis (Kao et al., 2016), systematic and cross-species characterization of this gene family across the broader Crustacea with focus on food crop species is currently lacking. A better understanding of homeobox genes in crustaceans is therefore required to address this major shortfall, leading us to our present work.
We retrieved complete transcriptome data sets for 120 crustacean species available at the time of manuscript preparation from the European Nucleotide Archive. Six non-crustacean arthropod proteomes were retrieved from Uniprot. A complete list of accessions used in this study is provided in Supplementary Table 1. We retrieved a list of query sequences used in subsequent homology searches from Uniprot and GenBank.
Based on a previously published workflow (Lai & Aboobaker, 2017), we used multiple Basic Local Alignment Search Tool (BLAST)-based approaches, such as BLASTp and tBLASTn to identify genes with homeodomain sequences. The BLAST results were filtered by e-value of < 10-6, best reciprocal BLAST hits against the GenBank non-redundant (nr) database and redundant contigs having at least 95% identity were collapsed using CD-HIT. We then utilized HMMER (version 3.1) employing hidden Markov models (HMM) profiles (Finn et al., 2011) to scan for the presence of Pfam homeodomains (Bateman et al., 2004) on the best reciprocal nr BLAST hits, to compile a final non-redundant set of crustacean and arthropod homeobox gene orthologs (Dataset 1).
Multiple sequence alignment of homeodomain sequences was performed using MAFFT (version 7) (Katoh et al., 2009). Phylogenetic tree was built from the MAFFT alignment using RAxML WAG + G model to generate a best-scoring maximum likelihood tree (Stamatakis, 2014). Geneious (version 7) was used to generate a graphical representation of Newick tree (Kearse et al., 2012).
With the recent availability of a large number of transcriptome data sets, we perform an extensive search for homeobox genes from 120 crustacean species. We focus on species represented across the broader Crustacea sampling from three main crustacean classes, Malacostraca, Branchiopoda and Copepoda, with focus on key food crop species from the order Decapoda (Supplementary Table 1). Using BLAST-based approaches and profile HMM (Bateman et al., 2004; Finn et al., 2011; Finn et al., 2015) for homology searches, we conservatively identified 4183 transcripts with homeodomain sequences from crustaceans (Figure 1; Dataset 1). Additionally, we included six non-crustacean arthropod species in our search and from these species, we identified 717 homeobox orthologs (Figure 1; Dataset 1).
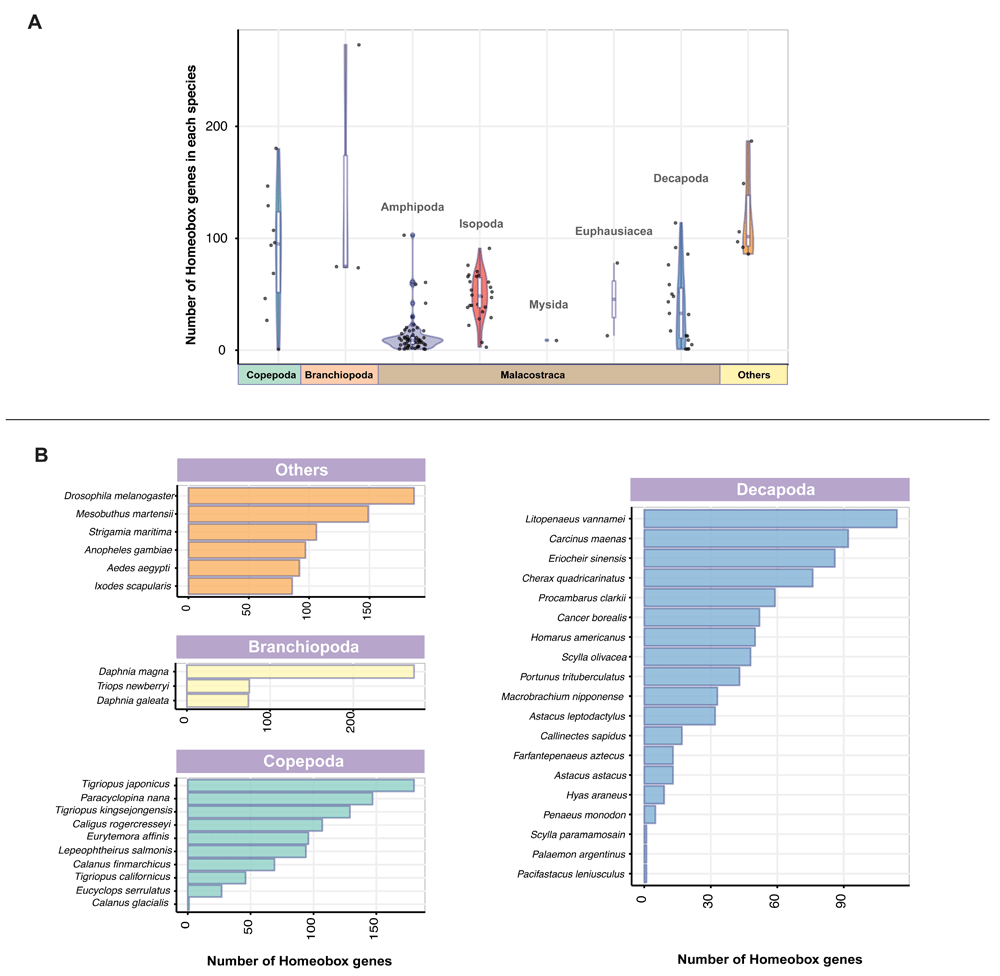
(A) Number of homeobox gene orthologs identified in each species are depicted as boxplots, indicating the median and quartiles. Violin plots underlying the boxplots illustrate sample distribution across different crustacean taxa and kernel probability density (width of the shaded areas represent the proportion of data located in these areas). The homeobox gene orthologs from six non-crustacean species within Arthropoda (others) are also shown. (B) Bar charts illustrating the number of homeobox gene orthologs in crustaceans from Decapoda, Branchiopoda and Copepoda along with six non-crustacean arthropods (others).
Concerted efforts to establish evolutionary classification of homeobox genes have resulted in 11 recognised classes (Edvardsen et al., 2005; Holland et al., 2007; Ryan et al., 2006; Zhong et al., 2008; Zhong & Holland, 2011). The Three-Amino acid-Loop Extension (TALE) superclass within the group of homeobox genes is characterized by three additional residues between alpha helices 1 and 2 of the homeodomain (Bertolino et al., 1995). TALE class homeodomain proteins are further divided into 6 subclasses, Meis, Pknox, Pbc, Irx, Mkx and Tgif characterized by distinct motifs beyond the homeodomain (Bürglin, 1997; Bürglin, 2005; Holland et al., 2007; Mukherjee & Bürglin, 2007). We have classified a total of 165 TALE class orthologs from 15 decapod crustacean species (Figure 2). These genes form distinct phylogenetic grouping, which allows confident assignment of decapod TALE class orthologs into 6 sub-families (Figure 2). Importantly, the tree topology of crustacean TALE class orthologs recapitulated observations from a previous study (Holland et al., 2007).
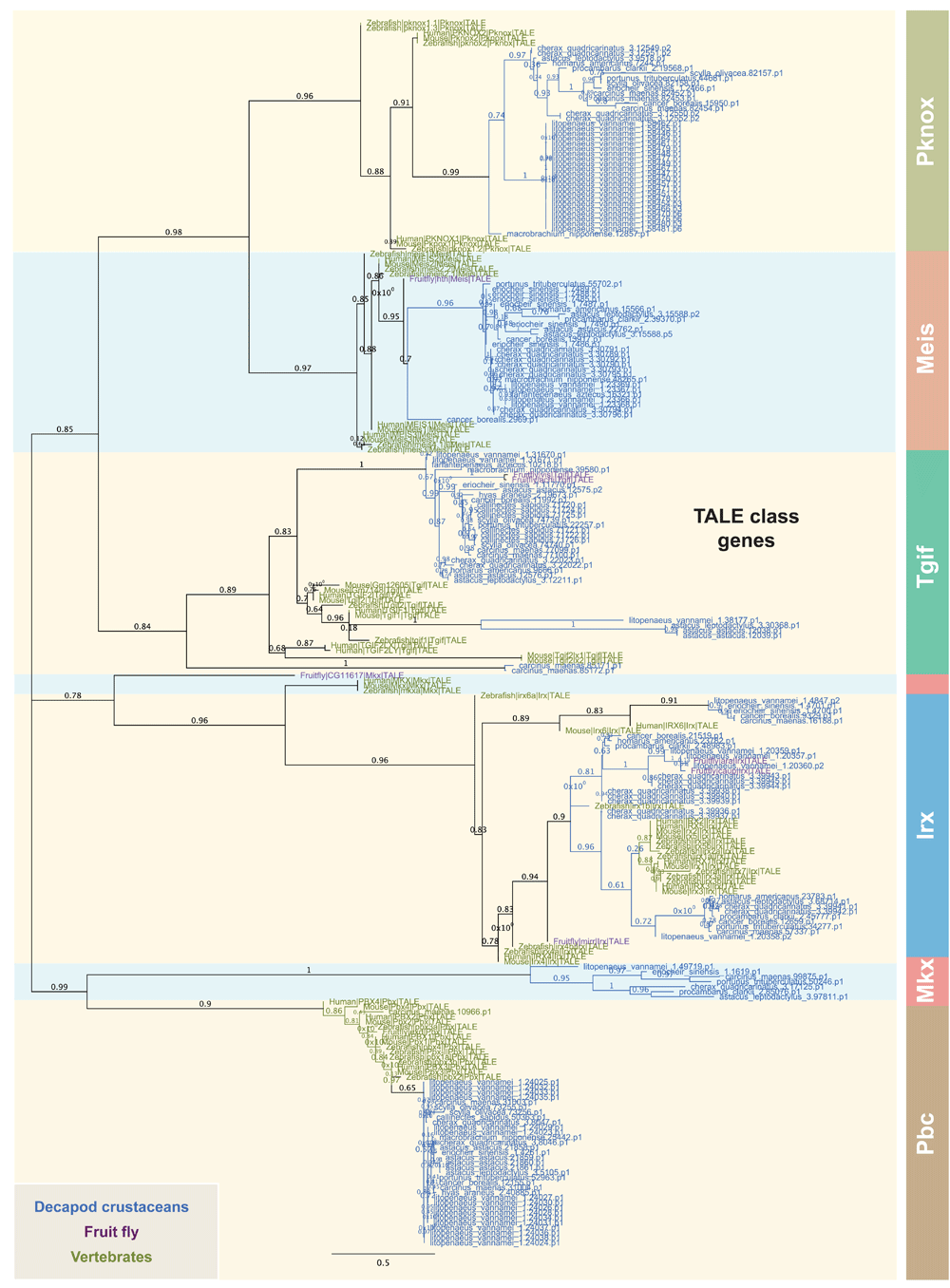
The tree was constructed using the maximum-likelihood method from an amino acid multiple sequence alignment, which include TALE class genes from other species (Zhong et al., 2008 and Zhong & Holland, 2011). TALE orthologs representing 6 subclasses are colour-coded. The node labels of each taxon are marked with distinctive colors denoted in the figure inset. Bootstrap support values (n=1000) are denoted as branch labels.
We identified 4900 homeodomain transcripts from 120 crustaceans and 6 non-crustacean arthropod species. Although this data set is non-exhaustive – transcriptomes contain only genes expressed at the point of sample collection – it will now serve as a key resource for future functional studies in the context of crustacean aquaculture. Beyond crustaceans, this work is widely applicable to studies on homeobox genes from other animals and will facilitate evolutionary and comparative genomics investigations.
Dataset 1: List of Pfam annotated homeobox genes and associated e-values in crustaceans and other arthropods. DOI, 10.5256/f1000research.13636.d190417 (Chang & Lai, 2018).
Dataset 2: Fasta file for homeobox gene sequences in crustaceans and other arthropods. DOI, 10.5256/f1000research.13636.d190418 (Chang & Lai, 2018).
This work was supported by the EMBO Fellowship and the Human Frontier Science Program Fellowship to AGL.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Supplementary Table 1: List of accession numbers for species used in this study.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Partly
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Transcriptional networks
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
Partly
If applicable, is the statistical analysis and its interpretation appropriate?
Not applicable
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |||
|---|---|---|---|
| 1 | 2 | 3 | |
|
Version 1 17 Jan 18 |
read | read | read |
Click here to access the data.
Spreadsheet data files may not format correctly if your computer is using different default delimiters (symbols used to separate values into separate cells) - a spreadsheet created in one region is sometimes misinterpreted by computers in other regions. You can change the regional settings on your computer so that the spreadsheet can be interpreted correctly.
Click here to access the data.
Spreadsheet data files may not format correctly if your computer is using different default delimiters (symbols used to separate values into separate cells) - a spreadsheet created in one region is sometimes misinterpreted by computers in other regions. You can change the regional settings on your computer so that the spreadsheet can be interpreted correctly.
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)