Keywords
olfaction, cell biology, atf5, vomeronasal organ, olfactory receptor, vomeronasal receptor
olfaction, cell biology, atf5, vomeronasal organ, olfactory receptor, vomeronasal receptor
Mice possess two olfactory organs: the main olfactory epithelium (MOE), which houses olfactory sensory neurons (OSNs), and the vomeronasal organ (VNO), which houses vomeronasal sensory neurons (VSNs)1. The MOE is thought largely to function in the detection of odors with no prior or innate behavioral importance, though there are notable exceptions2–5. The VNO, on the other hand, detects pheromones, which drive important social and reproductive behaviors6,7.
The VNO is a bilobal, cresecent-shaped neuroepithelium. It is neurogenic, giving rise to new VSNs throughout the life of the animal1,8. Immature VSNs are located at the tissue margins, or the tips of the crescents, while mature VSNs occupy more central areas. VSNs can be initially divided into two types, based on the expression of their primary signaling G proteins. Apical VSNs express the G protein Gnai2, as well as type I vomeronasal receptors (V1Rs). Basal VSNs express the G protein Gnao and type II vomeronasal receptors (V2Rs)9,10. V1Rs are expressed monogenically and monoallelically6,11,12. The situation is markedly more complicated for V2Rs. Type II VSNs express two V2Rs in non-random combinations: one from V2R family A, B, or D; and at least one from V2R family C13,14. In addition, some basal VSNs also express at least one gene from the non-classical MHC H2-Mv gene family15,16.
The VR(s) expressed by a VSN both drive its pattern of connectivity to the accessory olfactory bulb and define its receptive field12. Therefore, the choice of receptor(s) to express is considered to be a central gene regulatory decision in VSN development. It is thought that the V1R expressed by a given type I VSN is chosen stochastically during development1. In the case of type II VSNs expressing multiple V2R genes, given that the coexpressed receptors (and H2-Mv genes for basal type II VSNs) occur in non-random combinations, it is possible that the initial V2R choice event is stochastic but acts to restrict subsequent V2R or H2-Mv choice events.
The past decade has seen the discovery of many molecular players involved in the establishment of monogenic OR expression. Monogenic OR expression begins with OR gene choice, a complex process involving condensation of OSN chromatin17, extensive modification of the OR gene chromatin environment18,19, recruitment of cis and trans enhancer elements20,21, and cooperation between a number of transcriptional activators22,23. OR choice is followed by OR feedback, which functions to preclude further OR gene choice, to promote maturation of the OSN, and to stabilize expression of the chosen OR1,24–29,30. Together, OR choice and OR feedback ensure that each mature OSN expresses exactly one OR allele, defining each OSN as a sensitive and unambiguous signaling unit.
It was recently shown that ORs drive feedback by activating the unfolded protein response (UPR), a ubiquitous signaling pathway that homeostatically maintains the ER folding environment by modifying both its folding load and its folding capacity24,31. OR expression activates the ER-resident kinase PERK, which then drives phosphorylation of the translation initiation factor eif2α, resulting in global attenuation of mRNA translation initiation and a specific increase in translation of mRNA encoding the transcription factor Atf5. ATF5 is required for OSN maturation and expression of adenylyl cyclase 3 (AC3). AC3 expression suppresses activity of a histone demethylase required for OR choice, LSD1. If OR choice fails to drive Adcy3 expression, as is the case with some OR pseudogenes as well as Atf5 and Adcy3 mutants, expression of the chosen OR is extinguished. This phenomenon, termed ‘gene switching’27, appears to add a layer of quality control for ORs, and also indicates that OR choice is initially unstable. This lack of stability is probably due to the dual demethylase activities of LSD1, which presumably allow it to de-silence an OR allele, and then to re-silence the same allele19. In this model, LSD1 downregulation by AC3 is required for stable transcription of the chosen OR. Together, these data support a model in which OR feedback acts to promotes OSN maturation, to prevent further OR choice, and to stabilize expression of the chosen OR allele.
In contrast to the growing body of knowledge on OR gene regulation, comparatively little is known for VRs. However, a number of lines of evidence support a model in which both V1Rs and V2Rs employ a feedback signal similar to that used by ORs to prevent further VR gene activation. First, VSNs choosing a V1R pseudogene target axons widely across the accessory olfactory bulb, indicating that they have subsequently selected a second V1R gene. This finding suggests that V1R protein activates VR feedback. Second, VSNs that choose an OR gene knocked into a V1R gene locus do not express additional V1Rs. This result suggests both that canonical V1R signaling is unimportant—as ORs and V1Rs signal through different second messengers—and that ORs can activate VR feedback12. Third, heterologous V2R expression activates the UPR, and both V1Rs and V2Rs, like ORs, fail to traffick from the ER when expressed heterologously. V2R trafficking appears to involve replacement of the ubiquitous chaperone Calreticulin with a VNO-specific homolog, Calreticulin 4. For V1Rs, the mechanism of ER trafficking in VSNs has yet to be established, but does not appear to involve either Calreticulin 4 or the OR transporters Rtp1/232. Finally, it was recently shown that Atf5 is required for maturation and survival of basal VSNs. This study also showed that while Atf5 mRNA expression is ubiquitous in VSNs, ATF5 protein is expressed in more limited patterns, suggesting that Atf5 mRNA is under translational control in the VNO33. In sum, these data suggest that OR and VR feedback may employ a common framework, converging on PERK-driven translation of ATF5.
In order to begin to define the mechanistic outline of VR feedback, I have assayed ATF5 protein expression in a series of mouse mutants previously employed in studies of OR feedback. I have found that in Lsd1 mutant VNOs, ATF5 protein is absent, establishing a common genetic requirement for Lsd1 in ATF5 translation in both the VNO and the MOE. Appearance of ATF5 also required both the ER-resident kinase PERK and phosphorylation of the translation initiation factor eif2α, suggesting that ER stress drives ATF5 translation in basal VSNs. Finally, in adult animals, ATF5 is widespread and found in anatomical areas corresponding to both immature and mature VSNs, suggesting that mature VSNs experience continued or spurious ER stress events. Together, these results support a model in which V1Rs and V2Rs both employ ER stress-mediated feedback, potentially with different requirements for ATF5 and subsequently with different transcriptional outcomes.
Lsd1 has previously been deleted from the olfactory placode by crossing animals carrying loxP-surrounded Lsd1 alleles to animals expressing Cre recominase under the control of the FoxG1 promoter. These conditional mutants lose expression of most OR genes, resulting in a failure to translate ATF5 and a failure of OSNs to reach maturity19. To test whether VSNs and OSNs share a genetic requirement for Lsd1 in the ATF5 expression, ATF5 immunoreactivity was assayed in control (FoxG1-Cre; Lsd1 fl/+) and mutant (FoxG1-Cre; Lsd1 flfl) VNO at embryonic day 18.5 (E18.5). A later analysis was not possible due to the perinatal lethality of this combination of alleles. As can be seen in Figure 1A–B, control animals exhibited robust ATF5 immunoreactivity in the VNO. As previously described, ATF5 expression was found to be widespread and heterogeneous from cell to cell. In contrast, Lsd1 mutants did not have observable ATF5 expression (Figure 1C–D). Consistent with previous findings showing that Atf5 is required for VSN maturation and survival, the VNO of the mutant animals was greatly reduced in size. Despite its decrease in size, the VNO was still readily identifiable through the use of a number of structural features, including the surrounding bone and mesenchyme structure, bilateral symmetry, position relative to the MOE, and the presence of a lumen of stereotypical shape, adjacent to an epithelium with a single layer of apical sustentacular cells. Together, these data indicate that in the VNO Lsd1 is required for ATF5 expression, and by extension for VSN maturation. On the basis of these data, I hypothesize that VR expression is under Lsd1 control, and that VR expression drives ATF5 translation. This hypothesis will be addressed in further detail in the discussion section.
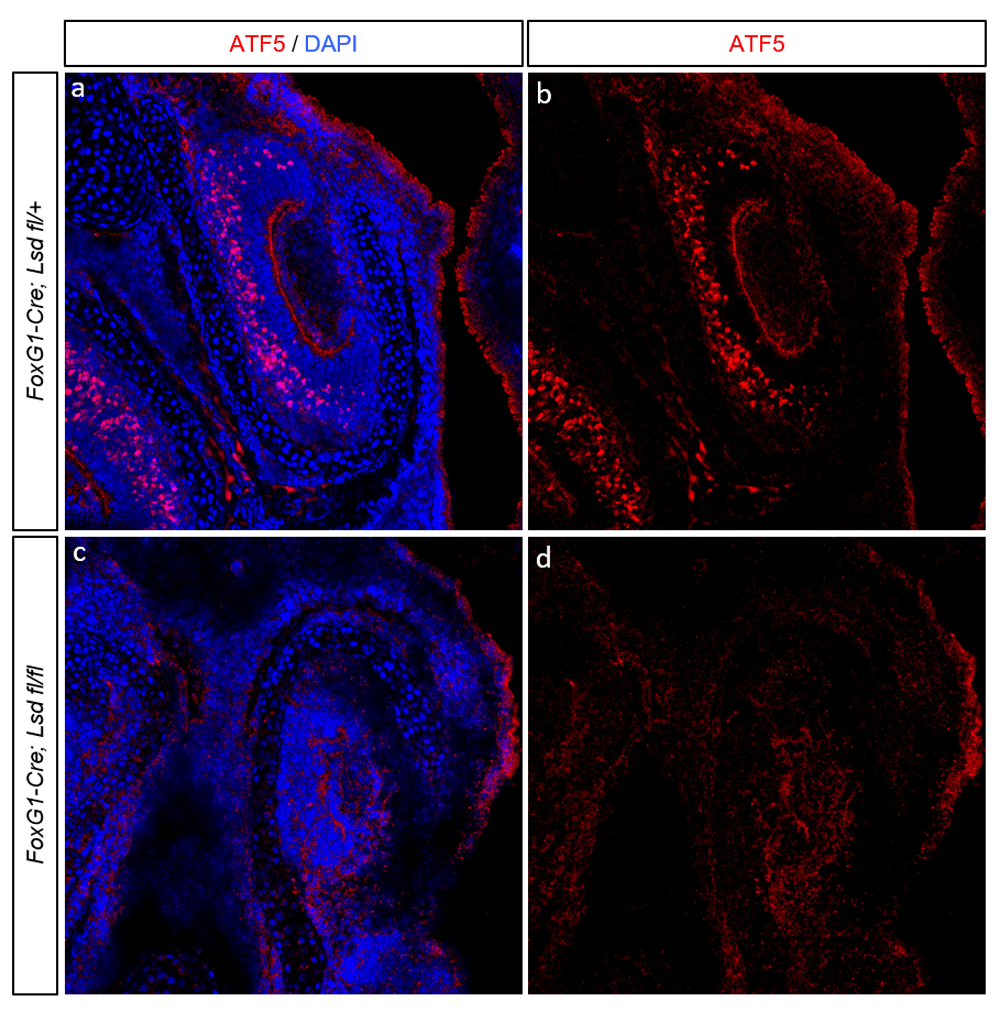
(A–B) Representative coronal section of embryonic day 18.5 (E18.5) VNO from a FoxG1-Cre; Lsd1fl/+ animal. (C–D) Representative coronal section from a FoxG1-Cre; Lsd1 fl/fl VNO, also stained for ATF5 and DAPI. For all images, ATF5 immunoreactivity is shown in red and DAPI nuclear counterstain in blue.
I next asked whether ATF5 translation in the VNO is under the same regulatory control as in the MOE. The Atf5 mRNA contains an inhibitory upstream open reading frame (iuORF) that under basal conditions suppresses its translation. However, upon phosphorylation of the translation initiation factor eIF2α at Serine-51, ribosomes bypass this iuORF to translate the Atf5 mRNA coding sequence24,34–37. OR expression in the MOE promotes this phosphorylation event and ATF5 translation by activating the ER-resident kinase PERK. OR-driven ATF5 translation can be blocked either through PERK deletion or through mutation of the serine phosphorylation site on eIF2α to alanine. I therefore asked whether ATF5 was lost in the VNO of PERK mutants and eIF2α phosphor-mutants. While P0 Perk+/- VNO exhibited robust ATF5 immunoreactivity (Figure 2A–B), ATF5 was completely absent in littermate Perk-/- animals (Figure 2C–D). Similarly, ATF5 was completely absent in eIF2αS51A/S51A animals, in which PERK is still present but cannot exert translational control through eif2α phosphorylation (Figure 2E–F). These data indicate that, as has been observed in the MOE and elsewhere, Atf5 mRNA in the VNO is under translational regulation via PERK-dependent phosphorylation of eIF2α.

(A–B) Coronal section of postnatal day 0 (P0) VNO from Perk+/- animal. (C–D) Coronal section of P0 VNO from a Perk-/- littermate. (E–F) Coronal section from a PO eIF2α S51A/S51A animal. For all images, ATF5 immunoreactivity is shown in red and DAPI nuclear counterstain in blue.
In the MOE, ATF5 expression is restricted to immature OSNs24. This expression pattern is intriguing, as both Atf5 and OR mRNA continue to be expressed in mature OSNs. It has been proposed that this context-dependence for ATF5 translation is due to increased expression of OR transporters such as RTP1/2 in mature OSNs, which could compete ORs away from PERK or simply relieve the ER burden imposed by ORs. In the VNO, a previous report demonstrated that at P0, Atf5 mRNA expression is essentially homogenous across the neuronal area, but that ATF5 protein is more heterogeneous. However, this report did not assay ATF5 expression in adult animals. While in young animals the VNO and MOE are dominated by immature neurons, in older animals the immature and mature neuronal compartments separate and resolve. In the adult VNO, a number of reports, using a variety of markers, have shown that immature VSNs are restricted to the VNO margins (i.e. the ‘tips’ of the VNO crescents)38,39. Surprisingly, ATF5 was not restricted to the tissue margins in the adult VNO. Instead, it was widespread, heterogeneous, and found in areas corresponding to both immature and mature VSNs (Figure 3A–B). Furthermore, co-staining sections from this animal with antibody against olfactory marker protein (OMP) to label mature VSNs revealed that some ATF5-labeled cells co-express OMP. While most OMP+ cells were either ATF5-negative, or displayed barely-detectable ATF5, other OMP+ cells displayed saturating levels of ATF5. These observations suggest that, unlike in the MOE, in VSNs ATF5 continues to be expressed after maturation. Given the nature of ATF5 translational control and the consequences of persistent UPR activation, this raises a number of interesting questions regarding PERK activation dynamics and the transcriptional output of ATF5 in VSNs.
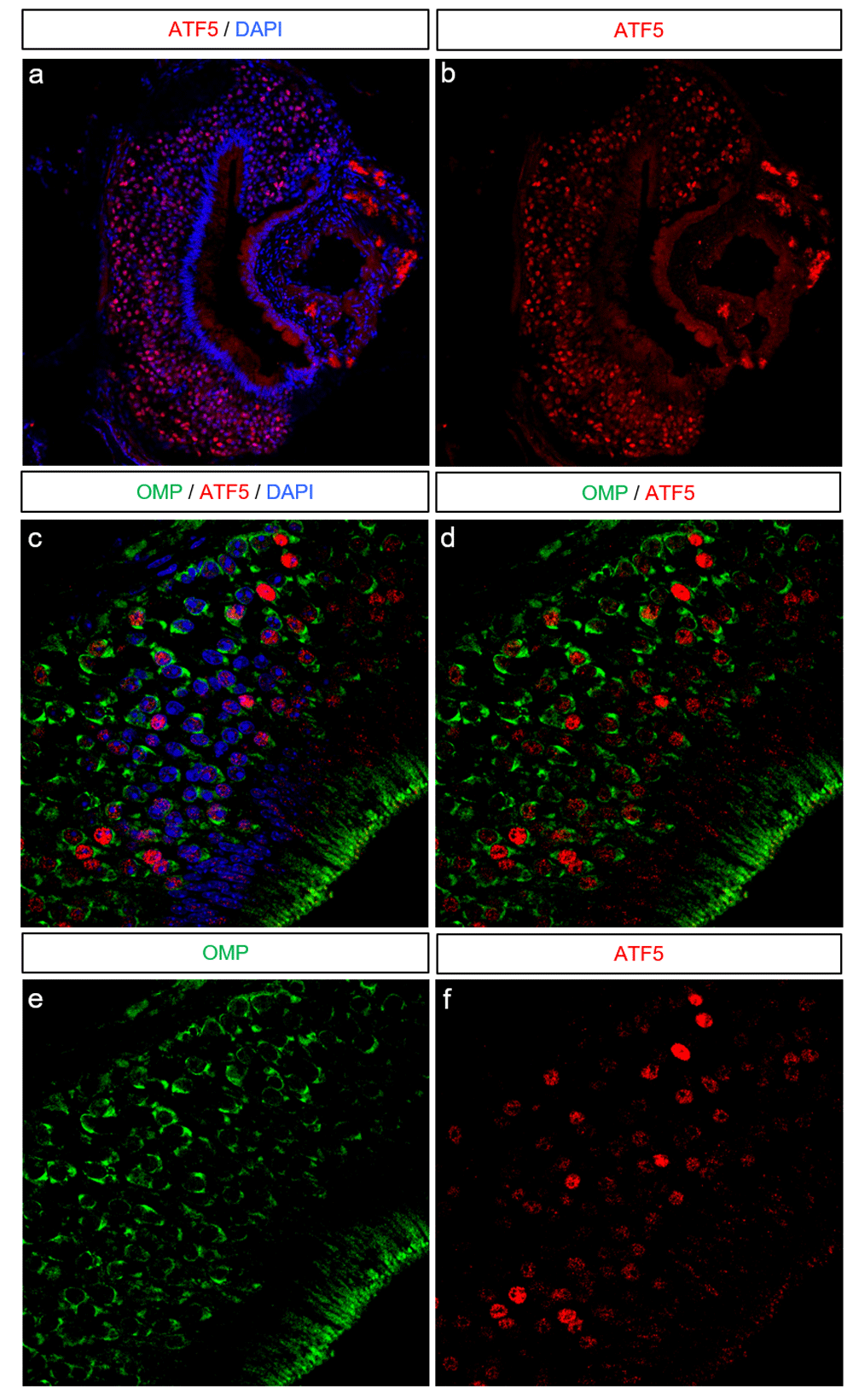
(A–B) Coronal section of a postnatal day 35 VNO. ATF5 immunoreactivity is in red and DAPI in blue. (C–F) A close-up section from the same VNO as shown in A–B. Olfactory marker protein (OMP) immunoreactivity is shown in green, ATF5 in red, and DAPI in blue.
Receptor-driven feedback programs endow developing olfactory neurons with a means by which to establish distinct, unambiguous cell fates. These programs are therefore essential in the construction of the basic architecture of the olfactory system. Furthermore, because these feedback programs allow the appearance of a single protein to establish cell fate, they also act as an engine in neuronal diversification, forging a direct relationship between the number of chemoreceptor genes and the number of chemosensory cell fates.
My previous work uncovered that in OSNs, OR feedback is executed by co-option of the PERK branch of the unfolded protein response24. An obvious follow-up question to this work was whether this feedback mechanism was employed in other tissues in which sensory cells express single or small numbers of sensory receptors. In the present work, I demonstrate that expression of ATF5, which is required for OR feedback and for the maturation and survival of basal VSNs33, has the same genetic requirements in the VNO and the MOE. This work therefore strongly suggests that ORs and VRs have a shared mechanism of feedback, converging on activation of the PERK branch of the UPR. This work was undertaken at the tissue level, and therefore more detailed analyses will likely be required in order to determine the specific requirements of VSN subtypes. Below I discuss some of the caveats of this work, as well as interesting questions for future work.
First, I hypothesized above that VR expression is under Lsd1 control and is required for ATF5 expression. Several pieces of data prompted this hypothesis. Among them are the shared elements in VR and OR feedback discussed in the introduction such as their activation of the UPR in cell lines, as well as the requirement of Lsd1 for ATF5 expression in the VNO demonstrated herein. However, it has yet to be directly demonstrated whether and how Lsd1 influences VR expression, or whether VR expression in VSNs drives ATF5 translation. A number of experimental considerations make these analyses difficult. Among them are the prenatal lethality of Lsd1 mutants and the requirement of Atf5 for VSN survival19,33, which together result in exceedingly small amounts of tissue for analysis of VR expression or of the epigenetic landscape of the VR gene family. Additional genetic models would likely be useful in determining the role of Lsd1 in VR choice, and a combination of biochemical and genetic approaches would be powerful in the determination of the mechanisms by which chemoreceptors influence PERK activity.
Second, it has been shown that a key element of OR feedback is AC3-driven downregulation of LSD119. In the MOE, LSD1 downregulation both prevents further OR choice and acts to stabilize expression of the chosen OR. LSD1 downregulation therefore must be exquisitely timed. No analogous situation has yet been demonstrated for VSNs. It is worth noting that the requirements for VSNs are likely different than for OSNs. In particular, VSNs choosing VR pseudogenes continue to express them while also selecting another VR from a different VR gene cluster40. This finding indicates that VR choice may involve the permanent engagement of a single or limiting element in cis to a given VR cluster. VR feedback may therefore act to prevent further choice, but not to stabilize VR expression. Thus, if VSNs employ a mechanism similar to that of AC3 in OSNs, it may only act to terminate further VR choice, but not to stabilize VR choice. The mechanistic basis of this difference is a fascinating area for future study.
Third, the convergence on ATF5 in OSNs and VSNs prompts a number of questions on the role and transcriptional output of ATF5. For example, how could ATF5 control OR feedback in OSNs and VR feedback in VSNs? It seems likely, given that ATF5 is a bZip-family transcription factor, that ATF5 has different binding partners in different tissues. This model would allow for co-factors to tune the transcriptional specificity of ATF5, but would prevent their engagement until ATF5 has been translated. For example, in OSNs this may allow ORs to promote expression of RTP1/2 such that they can subsequently be targeted to the plasma membrane. In contrast, given that basal VSNs express non-random combinations of receptors and that the expression of these receptors is sequential, expression of one VR may drive ATF5 expression to aid in selection of a second VR (or an H2-Mv). The identity of these potential binding partners is a fascinating outstanding question and is likely to greatly aid in our understanding of chemoreceptor feedback programs.
Fourth, as demonstrated herein, ATF5 continues to be expressed in mature VSNs, unlike findings in OSNs. In addition, cell-to-cell levels of ATF5 appeared to be extremely variable, with signal nearly undetectable in most cells, but reaching saturation levels in other cells. This is a fascinating observation, as it would indicate that mature VSNs continue to experience ER stress events. Atf5 is ubiquitous in VSNs33 and the UPR-driven mRNA translation program is rapidly induced but brief. I therefore hypothesize that the ATF5 expression patterns I observed reflect transient ER stress events experienced by many or all VSNs. However, it is impossible to rule out an alternate scenario in which some cells (or even VSN sub-types) experience continuing ER stress while others do not experience ER stress at all. An additional implication of the prolonged ATF5 expression pattern in VSNs could be that VRs and ORs have different mechanisms of PERK activation, for example direct versus indirect. A number of studies support an indirect model of PERK activation by ORs, in which ORs activate PERK only in the absence of RTP1/2, but this question is unaddressed for VRs. In addition, it is intriguing that ATF5 could be continuously expressed in mature VSNs, as it would beg the question of how VSNs can differentiate between bona fide ER stress and this developmental signal. Whether ATF5 has direct anti-apoptotic functions in VSNs as has been observed in other cell types has yet to be determined.
Finally, these findings firmly establish that a pathway canonically thought to be involved in the detection and resolution of cellular stress responses is fundamental in the designation of cellular identity and in cell maturation. This not only begs a reassessment of the role of PERK signaling, but also suggests specific additional studies. Given that activation of PERK provides such a powerful means by which to coordinate receptor appearance to the cellular gene expression program, and given that a multitude of cell types are defined by their expression of one or a handful of receptors, it would be surprising if PERK were not involved in other receptor-driven feedback programs. Excellent candidates include somatosensory neurons expressing Mas-related GPR family members, taste receptor cells, and photoreceptor cells. Specific chaperone or transporter requirements for these different receptors would provide a simple and generalizable mode for receptors to activate PERK in order to drive global gene expression programs, whose outputs can then be tuned by the use of tissue or cell type-specific co-factors.
All mice were housed in standard conditions with a 12-hour light/dark cycle and access to food and water in a UCSF barrier facility. All mouse experiments as well as euthanasia were approved by and were in accordance with University of California, San Francisco Institutional Animal Care and Use Committee (IACUC) protocol as described previously18. Animals used in this study were under protocols held by the Lomvardas laboratory. Details on standard procedures including euthanasia can be found at the UCSF IACUC website (http://iacuc.ucsf.edu/Policies/awStandardProcedures.asp). Because all animals described in this study were only used for tissue collection, the relevant UCSF IACUC sections are those that deal with proper euthanasia. For all animals used in this study, animals were single or pair-bred (for animals harvested during pregnancy) or were group housed (for animals harvested as adults). For prenatal experiments, pregnancies were timed such that pregnant females and pups could be harvested at the time points listed in the study. The age at which tissue was collected for each experiment is indicated in the figure legends. For all tissue collected, animals were euthanized at the indicated time point using a CO2 flow rate designed to minimize suffering and per IACUC regulations. Following CO2 exposure, animals were secondarily euthanized either by cervical dislocation for adults or decapitation for prenatal and perinatal pups, and then immediately dissected. Main olfactory epithelia or vomeronasal organs were dissected using clean, sterile, and recently sharpened tools.
All strains were maintained on a mixed (Black 6 / FVB) genetic background. The following mouse lines used in this study have been previously described: FoxG1-Cre; Lsd1 flox19, Perk and Eif2αS51A/S51A24. No sex-based differences in expression of the markers tested herein were identified in preliminary studies and the studies herein contained animals of both sexes. All efforts were made to minimize the number of animals used in performing this study.
Immunofluorescence (IF) was performed as previously described17,19,24. All animals were dissected immediately following euthanasia by CO2 exposure. Briefly, tissue was directly dissected into optimal cutting temperature compound (OCT, Tissue-Tek #4583). 14µm sections were air-dried on glass slides (VWR #48311-6703) for 10 minutes, fixed in 4% paraformaldehyde (PFA, Sigma #158127) in phosphate buffer solution (PBS) for 10 minutes, washed 3x5 minutes in PBS + .1% Triton-X (PBST), blocked for 1 hour in 4% donkey serum in PBST, then incubated with primary antibodies diluted in PBST and under coverslips (VWR # 470019-008) overnight at 4C. The following day, slides were washed 3x15 minutes in PBST and then incubated with secondary antibodies and DAPI in PBST at concentrations of 1:1000 under cover slides. Slides were then washed 3x15 minutes in PBST and mounted with vectashield (Vector Laboratories # H-1000) for imaging. Imaging was performed on Leica 700-series laser scanning confocal microscopes. The following antibodies were used: goat anti-Atf5 (Santa Cruz Biotechnology, SC-46934, dilution 1:250), rabbit anti-OMP (Abcam ab93127, dilution 1:250). For each panel, at least one mouse per genotype was sectioned. Differences were not noted between males and females. Mice were genotyped in-house using genotyping protocols suggested by the original generators of the mouse line. Genotyping protocols included positive and negative controls reactions. Full MOE or VNO were sectioned. To minimize variability between slides, control and experimental genotypes were sectioned onto the same slide. Slides with strong antibody signal and low background were selected for analysis. For each section, microscope settings were optimized for signal:noise. All image analysis was done in Fiji (version 2.0.0-rc-39/1.50b, build 8dcf1e65a6) and consisted only of changing brightness and contrast for each channel.
All efforts were made to ameliorate the suffering of animals used in this study. Animals received regular care from the author and from the animal facility personnel, as well as monitoring for health and injury. Unhealthy or injured animals were either treated or, in severe cases, humanely euthanized. In accordance with UCSF IACUC guidelines (see above), all animals were euthanized by exposure to CO2 at a rate deemed to minimize stress and suffering. Animals were not bred unnecessarily, and when possible multiple types of tissue were dissected from each animal and saved for potential future use.
Dataset 1: Raw images of immunofluorescence presented in figures 10.5256/f1000research.13659.d19056141
This work was funded by a Genentech Predoctoral Fellowship
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Not applicable
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Not applicable
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
No
Competing Interests: No competing interests were disclosed.
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Not applicable
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Partly
References
1. Nakano H, Iida Y, Suzuki M, Aoki M, et al.: Activating transcription factor 5 (ATF5) is essential for the maturation and survival of mouse basal vomeronasal sensory neurons.Cell Tissue Res. 2016; 363 (3): 621-33 PubMed Abstract | Publisher Full TextCompeting Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |||
|---|---|---|---|
| 1 | 2 | 3 | |
|
Version 1 17 Jan 18 |
read | read | read |
Click here to access the data.
Spreadsheet data files may not format correctly if your computer is using different default delimiters (symbols used to separate values into separate cells) - a spreadsheet created in one region is sometimes misinterpreted by computers in other regions. You can change the regional settings on your computer so that the spreadsheet can be interpreted correctly.
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)