Keywords
Pollen, Sperm, Cis-Natural antisense transcript, Arabidopsis
Pollen, Sperm, Cis-Natural antisense transcript, Arabidopsis
Natural antisense transcripts (NATs) are endogenous transcripts that contain sequences complementary to each other. NATs have been shown to regulate gene expression by generating small RNAs from the overlapping region (Zhang et al., 2013). NATs are classified into two subgroups according to the site of their biogenesis: trans-NATs and cis-NATs. Trans-NATs are transcribed from different genomic loci, whereas cis-NATs are transcribed from opposite strands of adjacent genes (Jin et al., 2008). Based on the relative orientation and overlap degree of two transcripts, cis-NATs can be categorized into three types: head-to-head (5′ to 5′), tail-to-tail (3′ to 3′) and fully overlapping (Jin et al., 2008). cis-NATs are widely present in plants, animals and fungi (Zhang et al., 2013). In plants, cis-NATs are important for pathogen resistance (Katiyar-Agarwal et al., 2006), stress tolerance (Borsani et al., 2005), successful fertilization (Ron et al., 2010), and phosphate homeostasis and plant fitness (Jabnoune et al., 2013).
Several genome-wide investigations identified potential cis-NATs in plants, ranging from 1057 to 1710 pairs in Arabidopsis (Henz et al., 2007; Jin et al., 2008; Wang et al., 2005; Zhan & Lukens 2013), and 3819 pairs in rice (Lu et al., 2012). However, all the expression data used in these cis-NAT investigations were from bulk samples such as seedlings, leaves or inflorescences, which include many cell types. For potential cis-NATs to be functionally relevant, the reverse and complementary transcripts must be co-expressed in the same cell. Some potential cis-NATs identified in those investigations might be expressed in different cells and, thus, the presence of overlapping transcripts in the same cell is not known. Moreover, the regions predicted to overlap in previous investigations were based on available gene model annotations, which might not fully represent potential overlaps, due to alternative splicing at different developmental stages or, to more extensive 5’ or 3’ UTRs than are annotated.
A pollen grain contains only two types of cell, one vegetative cell and two sperm cells, and thus can be used to investigate cell-specific cis-NAT pairs. Pollen RNA-seq data (Loraine et al., 2013) provides accurate transcript lengths, helpful for precisely identifying overlapping regions of two adjacent genes. Sperm microarray data (Borges et al., 2008) is helpful for defining whether two adjacent genes are expressed in the same cell. Moreover, a small RNA database for pollen and sperm (Slotkin et al., 2009) can be used to determine if small RNAs were detected from any overlapping regions.
In this study, we investigated potential cis-NATs in Arabidopsis sperm and vegetative cells using pollen RNA-seq data, sperm microarray data and sRNA data in pollen and sperm. In total, we identified 1471 cis-NAT pairs, including 131 novel pairs, with 72 and 56 pairs being potentially functional cis-NATs in sperm and vegetative cells respectively. These cis-NATs are tools for understanding gene regulation mechanisms in sperm and vegetative cells.
We investigated potential cis-NATs from protein-coding genes in pollen and sperm using pollen RNA-seq data (Borges et al., 2008; Loraine et al., 2013) and TAIR10 gene models (TAIR10 gene annotation data available here) using the Integrated Genome Browser, available from http://www.bioviz.org (Freese et al., 2016). We loaded TAIR10 gene models s and pollen RNA-seq into IGB, then manually scanned for cis-NATs in each chromosome, based on the following parameters: 1) the orientation of two adjacent genes in TAIR10 was reverse and complementary; 2) the length of transcripts mapping to the overlap of two adjacent genes was larger than 21nt, because the size of sRNA generated by cis-NATs is normally larger than 21 nt; and 3) both adjacent genes encoded proteins. The expression and sRNA data of each cis-NAT was merged with the cis-NAT data in Excel to generate sheet 1 of Table S1. Different categories of cis-NATs in sheets 2–8 of Table S1 were obtained based on the cis-NAT data in sheet 1 of Table S1.
We identified 1471 potential cis-NAT pairs, comprising 1373 pairs whose transcripts were complementary at their 3’ ends, and 98 pairs whose transcripts were complementary at their 5’ ends (Figure 1A, sheet 1 and 2 of Table S1). Among these 1471, in 37 pairs one transcript was completely internal to the other, 100 pairs comprised 50 sets that had three overlapping genes (sheet 1 of Table S1), and 131 pairs (8.9%) were not apparent using the TAIR 10 gene models, but were detected in the pollen RNA-seq data (Figure 1B and G, sheet 1 of Table S1).
One criterion for functional cis-NATs is that the two adjacent genes of a cis-NAT pair are expressed. To identify potentially functional cis-NATs in pollen, we analyzed the expression level of the 1471 gene pairs, defining genes with reads per million (RPM) ≥1 as expressed. There were 599 pairs for which the RPM of two adjacent genes was lower than 1 (sheet 3 of Table S1), suggesting that those 599 pairs might not produce relevant cis-NATs in pollen. Most sperm-specific genes, such as GEX1, GEX2 (Rutley & Twell, 2015), and Kokopelli (Ron et al., 2010), are detectable in pollen RNA-seq data, but sperm-specific genes with lower expression levels, such as ARI14, might not be detected, as the proportion of RNA from the vegetative cell is much larger than that from the sperm cells. So potential cis-NATs with one expressed gene in pollen RNA-seq data might still be functional in pollen. Based on this, there were 872 possibly functional cis-NATs pairs, for which the RPM of either or both adjacent genes was ≥1 (Figure 1C, sheet 4 of Table S1), of which 62 pairs did not overlap in TAIR10 gene models. Note that we did not detect a cis-NAT pair between Kokopelli and ARI14 (Ron et al., 2010), because the TAIR 10 gene model does not show them overlapping, and ARI14 expression is low in wild type. Thus it is possible that other cis-NATs might similarly not be included in sheet 4 of Table S1 (see below).
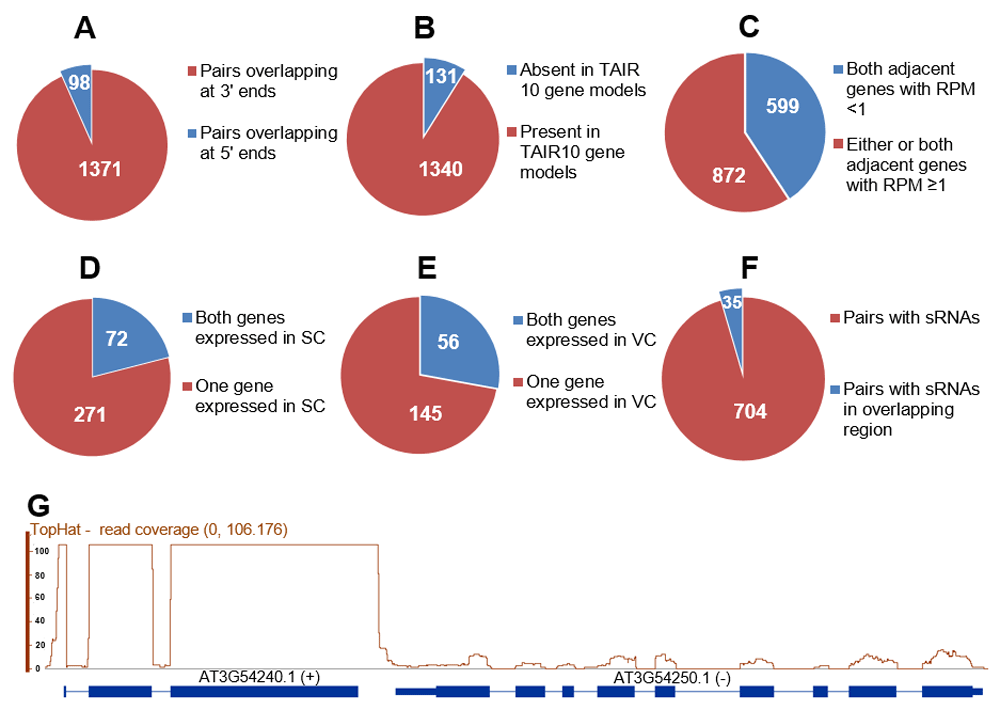
(A) Overview of potential cis-NATs with different overlapping types. (B) Overview of potential cis-NATs present or non-present in TAIR 10 gene models. (C) Overview of potential cis-NATs based on reads per million (RPM) from pollen RNA-seq data. (D) Overview of potential cis-NATs possibly functional in sperm cells (SC). (E) Overview of potential cis-NATs possibly functional in the vegetative cell (VC). (F) Overview of potential cis-NATs with sRNAs in vegetative and sperm cells.(G) Potential cis-NATs not present in TAIR10 gene models, but detected in pollen RNA-seq data.
Another criterion for functional cis-NATs is that both adjacent genes are expressed in the same cell. In order to accurately identify potentially functional cis-NATs in pollen, we investigated the gene expression level of 872 pairs in sperm cells. The microarray signal calls “Present (P) or Absent (A)” (Borges et al., 2008) were used to categorize genes as expressed or not expressed, respectively. We identified 72 pairs for which both adjacent genes were expressed in sperm, supporting the likelihood that these 72 cis-NATs pairs exist in sperm (sheet 5 of Table S1). There were an additional 271 pairs for which only one of the adjacent genes was expressed in sperm (Figure 1D and sheet 6 of Table S1), some of which might pertain to the Kokopelli/ARI14 example. To test if these pairs might function in the vegetative cell, we defined the gene as expressed in the vegetative cell if either 1): both pollen and sperm signals were called “P”, and the ratio of the pollen to sperm signal was > 3; or 2): the pollen signal was “P” and sperm signal was “A”. This exercise yielded 56 pairs for which both adjacent genes were expressed in the vegetative cell (sheet 7 of Table S1), and 145 pairs for which only one was expressed in the vegetative cell (Figure 1E and sheet 8 of Table S1). Another hallmark of functional cis-NATs is that there are small RNAs generated from the overlapping region. We therefore investigated the sRNAs of pollen and sperm (Slotkin et al., 2009) for these potentially functional cis-NAT pairs. sRNAs were detected at 794 genes, belonging to 739 pairs (Figure 1F and sheet 1 of Table S1). Of these, 35 cis-NATs pairs had sRNAs from the overlapping region.
cis-NATs are widely present in plants, and play an important role in regulating gene expression. However, in plants precise identification of cis-NATs at a cell-specific level to support whether cis-NATs might be functional is lacking. One possible reason was that it is difficult to get specific cell types for RNA-seq. As pollen grains contain only two types of cell, they are an excellent model to investigate cell-specific cis-NATs.
The vegetative cell forms a pollen tube that transports two sperm cells into the ovule for fertilization. Successful fertilization needs proper gene regulation in both the vegetative and sperm cells (Ron et al., 2010). The precise identification of cis-NATs in pollen is a tool for understanding the molecular mechanism of pollen tube growth and fertilization. The 131 novel potential cis-NATs in pollen, and the 72 and 56 potentially functional cis-NATs in sperm and the vegetative cell, respectively, provide candidates toward further uncovering the regulatory mechanisms of gene expression in the sperm and vegetative cells.
We identified 1471 potential cis-NAT pairs, including 131 pairs only detected in the pollen RNA-seq data. There were 872 pairs expressed in the same cell and thus possibly functional in pollen, while 72 and 56 pairs were potentially functional in sperm and vegetative cell, respectively.
Arabidopsis pollen RNA-seq alignments data were loaded into Integrated Genome Browser from the IGB Quickload site
Unprocessed sequence data are available from the Sequence Read Archive under accession SRP022162 (Loraine et al., 2013).
The pollen and sperm microarray data was from (Borges et al., 2008) (available at https://bmcplantbiol.biomedcentral.com/articles/10.1186/1471-2229-9-87)
sRNAs in Arabidopsis pollen and sperm were downloaded from https://mpss.danforthcenter.org/dbs/index.php?SITE=at_sRNA&lib_type=sRNA&lib_id=487 and https://mpss.danforthcenter.org/dbs/index.php?SITE=at_sRNA&lib_type=sRNA&lib_id=489, respectively (Slotkin et al., 2009).
This work was supported by United States Department of Agriculture Current Research Information System (5335- 21000-030-00D) funding to S.M.
Table S1. Investigation of cis-NATs in pollen and sperm cells of Arabidopsis.
Click here to access the data.
Sheet 1) List of all potential cis-NATs identified using TAIR 10 gene models and pollen RNA-seq data; Sheet 2) List of potential cis-NAT pairs with three overlapping genes; Sheet 3) List of potential cis-NATs pairs for which the RPM of both adjacent genes is < 1; Sheet 4) List of potential cis-NATs pairs for which the RPM of either or both adjacent genes is ≥ 1; Sheet 5) List of potential cis-NAT pairs for which both adjacent genes are expressed in sperm cells; Sheet 6) List of potential cis-NAT pairs for which only one gene is expressed in sperm cells; Sheet 7) List of potential cis-NAT pairs for which both adjacent genes are expressed in the vegetative cell; Sheet 8) List of potential cis-NAT pairs for which only one gene is expressed in the vegetative cell.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Epigenetics and small RNA in plants
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |||
|---|---|---|---|
| 1 | 2 | 3 | |
|
Version 1 22 Jan 18 |
read | read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)