Keywords
p53 Gene mutation, Codon 249, Hepatocellular carcinoma, p53 Exon 4, p53 exon 7
Human TP53 is the gatekeeper for generation of human cells and is highly conserved. Some alteration/mutation in TP53 adversely affects the regulatory function of the protein, potentially resulting in cancer. This study investigated mutations in codons 72 and 249 of TP53, among patients with hepatocellular carcinoma (HCC) and chronic hepatitis B virus (HBV) infection at the Moi Teaching and Referral Hospital (MTRH), Eldoret, Kenya.
In total, 33 HBV-positive patients attending MTRH hospital between September 2013 and July 2017 were purposely selected from medical records for the study; those with HCC were confirmed from the cancer registry. The patients were aged between 25-67 years, with a male-to-female ratio of 1.1:1. Blood samples were collected from the patients. DNA was extracted, amplified and sequenced using TP53 forward and reverse primers. Gene mutation detection and analysis was done on exons 4 codon 72 and exon 7 codon 249.
Of the 33 patients, 75.8% were chronically infected with HBV and had HCC; the rest were HBsAg positive without HCC. Homozygous proline was prevalent (54.5%) at exon 4 codon 72, followed by heterozygous Arg/Pro (33.3%) and lastly homozygous Arg/Arg (12.1%). Pro/Pro allele was frequent in HCC group while Arg/Arg allele was common in patients without HCC. There was no significant association between the HCC and codon polymorphisms (P=0.12). In exon 7, codon 249, 24.2% of patients had an Arg/Ser mutation of which, 75.0% had HCC and 25.0% did not. There was no significant association between HCC patients and codon 249 mutation (P=0.15).
TP53 is a gene gate keeper, the mutations under study may dependently play a role in HCC development. This study did not find any association between TP53 mutations and presence of HCC. Therefore, TP53 Arg-72 and Ser-249 mutation is not a clear prognosis indicator for hepatocellular carcinoma among HBV infected patients in Kenya.
p53 Gene mutation, Codon 249, Hepatocellular carcinoma, p53 Exon 4, p53 exon 7
The main reason for this version is to submit our responses
addressing comments raised by the reviewers (Lamech
Mwapagha and Harris Onywera) so that we complete the
publication process.
In this version, we have deleted information on exon 6 so that this
publication will concentrate on polymorphisms of exon 4 and 7.
Abstract: We have edited the conclusion to align with the
findings “TP53 Arg-72 and Ser-249 mutation is not a clear
prognosis indicator for hepatocellular carcinoma.”
In methods: We have provided the justification of conducting
the study at MTRH. All reagents and machines used have the
name, the manufacturer and town/city. In PCR amplification, we
have provided expected band size in the electrophoresis.
In Results: We have provided GenBank accession numbers
(MN119310-MN119350) of the sequences generated for
confirmation. We have provided Figure 2, which is a summary
of aligned TP53 exon 4, codon 72 amino acids sequence. This
figure will provide a quick visualization of the mutations. We have
provided additional information in caption of Figure 1 to explain
the details in the figure. Meaning of abbreviation used in Table 3
and Table 4 are now explained below each table.
In discussion we added a statement as one of the limitations of
the study as suggested by the reviewers.
In reference, we have added reference 9 (Hall et al., 2011) and
30 (Kumar et al., 2016).
To read any peer review reports and author responses for this article, follow the "read" links in the Open Peer Review table.
AFB1: Aflatoxin B1; BLAST: Basic Local Alignment Search Tool; CHB: Chronic Hepatitis B; DNA: Deoxyribonucleic acid; EDTA: Ethylene diamine tetra acetic acid; G: Guanine; HBsAg: Hepatitis B surface Antigen; HBV: Hepatitis B virus; HCC: Hepatocellular carcinoma; HCV: Hepatitis C virus; KEMRI: Kenya Medical Research Institute; MEGA: Molecular Evolutionary Genetics Analysis; IARC: International Agency for Research on Cancer; MTRH: Moi Teaching and Referral Hospital; NCBI: National Center for Biotechnology Information; PCR: Polymerase Chain Reaction; T: Thymine.
Hepatocellular carcinoma (HCC) is the fourth most common malignancy according to the World Health Organization (2022). HCC is increasing in incidence and has a mortality incidence of 800,000 deaths globally per year (Stewart et al., 2016). Reported incidences of HCC vary worldwide, with the West, Asia and Africa having the highest incidence rates. According to report on the Global Burden of Disease Cancer Collaboration et al. (2017) HCC is the fifth and seventh most common cancer in men and women, respectively. There are various causes of HCC, of which the most common is chronic infection with hepatitis B virus (HBV) and hepatitis C virus (HCV). In Kenya, those infected with HBV constitute 78.0% of HCC cases (Mutuma et al., 2011).
HCC is the primary liver cancer derived from uncontrolled multiplication of hepatocytes (Gomes et al., 2013). Just like in any other cancer, TP53 has a crucial role in HCC tumor suppression. The gene hampers progression of the cell cycle if DNA is damaged (Kruiswijk et al., 2015; Sasaki et al., 2011), a role that is inactivated in most cancers mainly through alteration in TP53, which can be caused by external agents (Gomes et al., 2013; Tokino & Nakamura, 2000). TP53 alterations are observed in most cancers and they affect major regulators of various signaling pathways involved in tumor suppression (Kandoth et al., 2013; Yang et al., 2013).
TP53 has ten coding exons, with mutations distributed in all of them, with a strong predominance in exons 4–9, encoding the DNA-binding domain of the protein (Rivlin et al., 2015). Studies have demonstrated that mutant TP53 contributes immensely to replication of damaged DNA and to tumor progression. These mutant proteins bind to TP53 response elements thereby weakening the process of DNA repair and TP53-mediated apoptosis (Carvajal et al., 2012; Maiuri et al., 2010). In exon 7, codon 249 (AGG→AGT, arginine to serine) has been identified as a “hotspot”. Differences in ethnicity and geographical location among other factors have varied impact on TP53 codon 249 (AGG→AGT) mutation profiles (Kandoth et al., 2013; Wen et al., 2016). In exon 4, an arginine to Proline substitution at codon 72 has been investigated as risk modifier in several cancer models; however, its role in cancer progression remains uncertain.
There is paucity of information on TP53 in Kenya. In this study, we evaluated the presence of TP53 gene mutations in exons 4 and 7 among HCC patients attending Moi Teaching and Referral Hospital (MTRH), in western region of Kenya.
The samples were collected from jaundiced patients chronically infected with HBV attending MTRH, Eldoret, Kenya between September 2013 and July 2017. The MTRH was selected as it is one of the largest national referral hospitals in western Kenya, where rates of HBV infection was found to be 50% among patient with jaundice (Ochwoto et al., 2016). A mixed method designs were used in this study. First, patients were purposively selected from hospital records based on them being jaundiced and HBV-positive with or without HCC. Secondly, the patients were contacted and then recruited in person. Lastly, those with HCC had their cancer status confirmed using the cancer registry of Eldoret Hospital, Uasin Gishu, Kenya. A patient with HCC was defined as having liver cancer based on the patient’s medical record and cancer registry file. All patients with HCC were selected. Other patients’ medical records obtained from the hospital included gender and residential area. The male-to-female ratio was 1.1:1 and the age range was from 25 to 67 years. None of the patients had received any viral HBV treatment by the time of sample collection.
The ethical approval to conduct the study was obtained from Institutional Research and Ethics Committee (IREC) of MTRH/Moi University (approval number 001002), from Kenya Medical Research Institute Scientific Ethics Review Unit (approval number KEMRI/SERU/CVR/001/3211) and from Eldoret Cancer Registry (approval number ECR/DRA/2017/001). Further, the participants had to provide informed consent for the study prior to blood draw.
Blood samples were collected in vials anti-coagulated with EDTA. Plasma was separated at MRTH and thereafter the plasma tubes were shipped on dry ice to the KEMRI Production Unit in Nairobi. The samples were then stored in aliquots at -80°C until subsequent testing.
Screening for hepatitis B virus surface antigen (HBsAg) and antibody to the core protein (anti-HBc) were performed using the COBAS e411 platform (Elecsys; Roche Diagnostics, Quebec, Canada). Chronic hepatitis B (CHB) was determined by anti-HBc IgM positive serology, as described previously (Park et al., 2015).
Circulating DNA of Human TP53 tumor suppressor gene was extracted from 200 µl of plasma samples using QIAmp DNA mini-extraction kit (Qiagen Inc, Germantown, Maryland, USA) according to manufacturer’s instructions. The DNA was subsequently eluted in 60 µl of AE buffer and quantity measured by NanoDrop spectrophotometer (Thermo Scientific, Wilmington, Delaware USA) and stored at -30°C until use.
Three different primers targeting TP53 gene exons 4 and 7 (Table 1) were used in amplification of the DNA extracts using conventional PCR. The PCR mix targeting the two exons was similar except for the primer. Each PCR tube contained a total volume of 50 µl reaction mixture, with 5 µl of 10 µg human genomic DNA template or AE buffer for negative controls, 5 µl of 10X PCR buffer 5 µl of 25 mM MgCl2, 5 µl of 1.25mM dNTP mix, 0.2 µl of 5U of Taq DNA polymerase (Qiagen Inc, Germantown, Maryland, USA), 1.25 µl each of a 20 uM stock of forward and reverse primer of sequences (Table 1).
| Primer | Sequence (5’ to 3’) | Expected band size |
|---|---|---|
| Exon 4 forward | ATCTACAGTCCCCCTTGCCG | 296bp |
| Exon 4 reverse | GCAACTGACCGTGCAAGTCA | |
| Exon 7 forward | CTTGCCACAGGTCTCCCCAA | 254bp |
| Exon 7 reverse | AGGGGTCAGCGGCAAGCAGA |
The mix was loaded to Veriti, 96 well thermal cycler machine (ABI systems Foster, California, USA). PCR amplification profile for exon 7 was set at 95°C for 10 minutes initial denaturation and 35 cycles of denaturation at 95°C for 45 seconds, annealing at 58°C for 30 seconds and extension at 72°C 30 seconds. Final extension was at 72°C for 10 minutes. For exon 4 the PCR amplification profile was set at 94°C for 12 minutes initial denaturation and 35 cycles of denaturation at 94°C for 40 seconds, annealing at 56°C for 30 seconds and extension at 72°C 30 seconds. Final extension was at 72°C for 10 minutes.
After that, a 4 µl aliquot of PCR product was electrophoresed by using 2% agarose (Fisher Scientific), 2 µl of 5X Gelpilot DNA loading Dye (Qiagen Inc, Germantown, Maryland, USA) together with 100 bp track DNA ladder (Invitrogen, California, USA) in 1X TBE buffer containing SYBR-safe DNA gel stain (Invitrogen, California, USA) and visualized using an ultraviolet trans-illuminator gel Doc-It2 Imager then viewed using Vision Works LS software v.7.1.
For exon 7, 5 μl of the all negative amplicons was used in the second nested PCR (forward primer exon 7b 5-AGGCGCACTGGCCTCCTT-3 and reverse primer exon 7b 5-TGTGCAGGGTGGCAAGTGGC-3). The master mix and the PCR profile of the nested PCR were similar to the first round profile. The amplicons were viewed following electrophoresis on a 2% agarose gel.
All PCR-positive amplicons were purified using the Qiagen Gel purification kit according to the manufacturers recommended protocol. The purified DNA was quantified using a Nanodrop spectrophotometer (Thermo Fisher Scientific), and purified DNA (50 ng) was sent for Sanger sequencing at Macrogen, Inc. (Netherlands) using the first primer sequences and the manufacturer’s guidelines
The directly amplified sequences were assembled using GENETYX version 9.1.0 (GENETYX Co., Tokyo, Japan; PCAP is an open-access alternative) DNA sequence analysis software. The sequences were aligned to TP53 gene sequences using NCBI BLAST for identity confirmation. The contigs from GENETYX were then aligned to the TP53 gene reference sequence from the International Agency for Research on Cancer (IARC) database using Bioedit software version 7.2.5 (Hall et al., 2011). Mutations to the sequences were analyzed using MEGA v.7.0 (Kumar et al., 2016) software.
Test for statistical significance of mutation profile parameters were done using the χ2 test and Fisher’s exact test. P-values less than 0.05 were considered statistically significant. To examine possible associations between mutations in TP53 exons and hepatocellular carcinogenesis, we analyzed 2x2 tables using Fisher’s exact test. Odds ratios (ORs) were used to analyze two significant associations at 95% confidence interval (CI). Statistical analysis was performed using SAS version 9.4.
There were 33 subjects in total for whom results in exon 4 and 7 were obtained. The characteristics of the subjects are shown in Table 2. The ratio of male to female was 51.5% to 48.5%. All the subjects were positive for HBsAg. Those who were chronically infected and had HCC were 75.8% (25/33), of which 48.0% were female and 52.0% were male. Among those that did not have HCC but were HBsAg positive (24.2%), half were female and the other half male.
A total of 33 samples were amplified with clear forward and reverse sequences for exon 4 codon 72. Amino acids of the aligned sequences (Figure 2) revealed that majority (54.5%) of polymor-phisms at codon 72 were Pro/Pro (CCC) alleles, followed by heterozygous Arg/Pro (33.3%) and homozygous Arg/Arg (CGC) (12.1%) (Figure 1). All those homozygous for Arg/Arg were male. The sequences generated from this study were deposited in GenBank with accession number MN119310-MN119350 and aligned in Figure 2 using MG595994.1 as reference sequence.
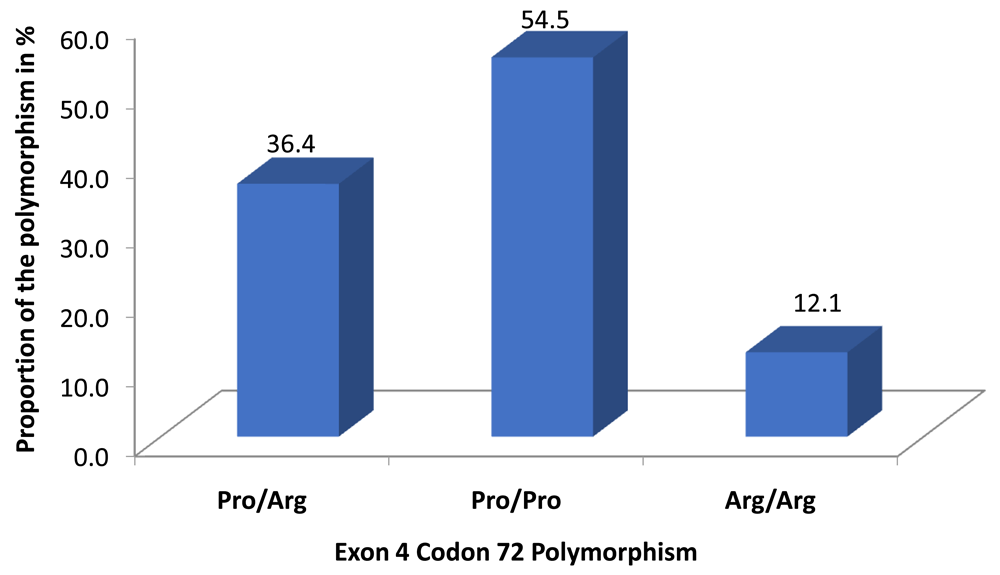
Majority of the patients had Proline amino acid at this codon followed by a mixture of Proline (Pro) and Arginine (Arg).

There was statistically significant association between the sex of the subject and the polymorphism identity (Fisher’s exact test=5.4 and P=0.04), with all the homozygous Arg/Arg belonging to male patients, whereas a higher proportion of female patients had homozygous Pro/Pro (64.7%) compared to male patients (35.3%) and a higher proportion of male had the heterozygous Pro/Arg(58.3%) compared to female patients (41.7%). On the other hand there was no statistical significance between the HCC and the polymorphisms (Fisher’s exact test=3.58 and P=0.12). However, it is important to note that at codon 72 most of the patients with HCC had Pro/Pro alleles, followed by heterozygous Pro/Arg and lastly homozygous Arg/Arg (Table 3). The Pro/Pro allele was more frequent in the HCC group, whereas all female patients with Arg/Arg alleles did not have HCC (Table 4).
There was no significant association between HCC, gender and TP53 codon 72 Pro/Arg when both HCC cases and the non-HCC cases were compared (P=0.57). Equally there was no significant association between HCC, gender and TP53 codon 72 Pro/Pro (P=0.40; Table 4)
Out of the 33 subjects, eight (24.2%) had the Arg>Ser codon 249 mutation and the majority (75.8%) did not have the mutation. Serine 249 mutation was seen more in males (87.5%) than females (12.5%) and there was an association between the sex and mutation (Fisher's exact test=5.47, P=0.04) with male at higher risk compared to female (OR=10.5, 95% CI =1.1-98.9%) (Table 5).
Majority (75.0%) of those with the serine 249 mutation had HCC; only (25.0% with the mutation were without HCC) (Table 5) Similarly, among those without the mutation, 76.0% had HCC and 6 (24.0%) did not have HCC. The findings showed no significant association in the presence of codon 249 mutations between patients with and without HCC (P=0.15) at 95% CI (OR=0.52: 95% CI 0.054-4.773).
The association between HCC and mutations at codon 72 or 249 of TP53 remains controversial. To our knowledge, this is the first information concerning TP53 exon 4 and 7 mutation in Kenya among HBV-positive patients with and without HCC. A number of studies have described two structurally different forms of wild-type p53 resulting from the substitution of a proline for an arginine at residue 72, with different biochemical and biological characteristics (Thomas et al., 1999). Different prevalence of this substitution has been reported in various studies. In our present study, the homozygous Pro/Pro genotype was the most common (54.5%), and the least is homozygous Arg/Arg. This prevalence of allele is similar to Taiwanese (Mah et al., 2011), Egyptian (Neamatallah et al., 2014) and Chinese (Wang et al., 1999). We observed that patients with HCC had higher frequencies of Pro/Pro (88.2%) compared to observation made among Moroccan population (11.8%) (Ezzikouri et al., 2010) and Egyptian patients with HCV (Koushik et al., 2004; Neamatallah et al., 2014).
The association between TP53 codon 72 Pro/Arg gene polymorphism and liver cancer remains controversial, with some studies showing and others lacking the association. Among the studies that show associations, Dong et al., 2018 found that the TP53 Pro allele and Pro/Pro genotype were associated with cancer risk (Dong et al., 2018). In Egyptian patients with HCC, development of HCC was associated with Pro/Pro allele carriage as compared to Arg/Arg or Arg/Pr alleles (Neamatallah et al., 2014). This study did not find any association between polymorphisms and HCC among the HBV-positive patients. Other studies comparing acute hepatitis C and HCC have not found any association between codon 72 polymorphism and disease severity or HCC (Eskander et al., 2014; Hu et al., 2014). The inconsistency in association and prevalence observed could be attributable to geographical, environmental or ethnic differences in the studied population.
Our findings show the presence of selective guanine-to-thymine transversion mutation in the third base of codon 249 of TP53 in DNA isolated from 24.2% HCC patients. This mutation corresponds to arginine-to-serine substitution. Our findings corroborate data available among populations from Guangxi, Taiwan and The Gambia, where similar mutations were reported (Mah et al., 2011; Özdemir et al., 2010). However, evidence presented from a European population, which reported no mutation, is contrary to our findings (Kirk et al., 2000). According to a report by the Global Burden of Disease Cancer Collaboration et al. (2017), differences in geographical location, ethnicity, hereditary disorders, and excessive exposure to mutation-inducing agents as well as study size population could explain the discordance observed in the reported findings between our study and European studies (Global Burden of Disease Cancer Collaboration et al., 2017). Our study found no significant association between codon 249 mutation and hepatocellular carcinogenesis. However, exposure to codon 249 mutation might be considered a predisposing factor for HCC (OR=0.5278; 95% CI 0.0584-4.7736). These findings are in agreement with finite data available in Taiwan, United states, Japan, Australia, Gambian and Guangxi populations (Kirk et al., 2000; Mah et al., 2011; Özdemir et al., 2010; Stern et al., 2001). Likewise, array of lit-erature is available implicating that the presence of this very mutation in HCC patients from developed countries including the United States, China, Japan and Australia is remarkably low (Bruix et al., 2011; Kirk et al., 2000; Özdemir et al., 2010).
We further found that males were overrepresented in the mutation positive categories in patients with and without HCC. This could be ascribed to possible occurrence of faster and more severe HCC in males than females (Li et al., 2017). However, there was an association between the sex and mutation (Fisher's exact test=5.47, P-value =0.04).
Counter-intuitively, TP53 codon 249 mutations were observed not only in HCC patients but also in the non-HCC patients, thus, corroborating earlier findings by Kirk et al. (2000) that reported codon 249 mutation presence in 3 of 53 control subjects (6%), and those of Ozturk (1995), who reported codon 249 mutations in non-malignant liver tissues. A possible explanatory analysis for this finding is that mutations to codon 249 is generally known as a hotspot for aflatoxin B1 (AFB1)-driven modification. According to Özdemir et al. (2010), AFB1 induces codon 249 mutation among cancer patients residing in AFB1 high-risk regions, where chronic HBV and HCV infections are also endemic. Furthermore, among TP53 mutations described in human cancers and compiled in the IARC TP53 mutation database, 66% occur in patients with HCC originating from regions with a high incidence of HCC and high exposure to dietary AFB1. However, we did not perform aflatoxin exposure tests for the subjects to corroborate this. Additionally, published data from the Ministry of Health and the Gastroenterology Society of Kenya on guidelines for the treatment of HBV and HCV infections in Kenya (2015) suggested that 80% of HCC cases in the country are due to chronic infection with HBV (Ochwoto et al., 2016). This evidence perhaps indicates that the existence of the mutation in TP53 may be suggestive of an early genetic event in hepatocellular carcinogenesis. Consequently, it is argued that presence of a single mutation alone in DNA is unlikely to cause cancer, rather cumulative or multiple mutations in tumor suppressor genes are required (Adjiri, 2017; Lv et al., 2013).
Although this study investigated for the presence of TP53 mutation in exon 4 and 7 hepatocellular carcinoma patients, there is a need to look at the remaining exons. The cross-sectional nature of the study limited our analysis. Thus, we were unable to perfume any analysis to determine the evolution of the TP53 mutations among the patients with HCC. The use of samples that only amplified the forward and reverse fragments of exon 7 and 4 could bias the mutational prevalence. The mutations reported in this study were found in samples taken from the patients’ blood and we did not obtain tumor tissues from the patients for verification. Secondly, the study involved HBV infected patients, using the acute infected patients as negative control population; we recommend that involvement of health participants who are not infected with HBV and may be the same ages and gender may result to deeper understanding of exon 4 and 7 mutations and their roles in HCC development.
TP53 is a gatekeeper gene, and codon 72 and 249 mutations could play a role in HCC development. However, this study did not find any association between patients infected HBV with or without HCC. We further did not observe any clear mutational patterns between TP53 mutations and HCC development. We therefore conclude that in a cross sectional set up, TP53 mutation is not a good indicator for prognosis or tumor marker among HBV positive subjects in Kenya.
We acknowledge the study participants and the staff of Moi Teaching and Referral Hospital.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Partly
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: clinical pathology , subspecialty is chemistry and molecular studies
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
Partly
If applicable, is the statistical analysis and its interpretation appropriate?
Partly
Are all the source data underlying the results available to ensure full reproducibility?
Partly
Are the conclusions drawn adequately supported by the results?
Partly
References
1. Eskander EF, Abd-Rabou AA, Yahya SM, El Sherbini A, et al.: "P53 codon 72 single base substitution in viral hepatitis C and hepatocarcinoma incidences".Indian J Clin Biochem. 2014; 29 (1): 3-7 PubMed Abstract | Publisher Full TextCompeting Interests: No competing interests were disclosed.
Reviewer Expertise: Microbiome, Sexually Transmitted Infections, and Cancer
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
Partly
If applicable, is the statistical analysis and its interpretation appropriate?
I cannot comment. A qualified statistician is required.
Are all the source data underlying the results available to ensure full reproducibility?
Partly
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Cancer Genomics
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |||
|---|---|---|---|
| 1 | 2 | 3 | |
|
Version 2 (revision) 12 Mar 24 |
read | ||
|
Version 1 06 Aug 19 |
read | read | |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)