Keywords
PIK3CA, Gene Amplifications, Ovarian carcinoma, Quantitative Real-Time PCR, Omdurman Maternity Hospital.
PIK3CA, Gene Amplifications, Ovarian carcinoma, Quantitative Real-Time PCR, Omdurman Maternity Hospital.
Universally, ovarian carcinoma (OC) is the deadliest gynecological tumor. It is ranted as the fifth most prevalent disease causing mortality in women1,2. In 2018, there were 14,070 associated deaths and 22,240 new cases reported in the USA3,4. In Sudan, OC ranked as the fourth most common malignancy in women with 188 in every 100,000 people recorded; 8 in every 100,000 people at a gender-specific rate and 7 in every 100,000 people at an age-standardized rate5,6. In addition, a common issue for OC is that more often a higher stage of carcinoma is seen at diagnosis; resulting in a poorer prognosis for this cancer and the 5-year survival rate of advanced-stage OC remains low7,8.
The phosphatidylinositol-3-kinases (PI3Ks) are a member of the kinase family9,10 and have regulatory functions in cell survival, proliferation, and migration11,12. They also have a crucial role in cellular tumorigenesis, cancer development, and drug conflicts13,14. Mechanisms for pathway activation in cancer are mostly due to PIK3Ca or AKT (serine/ threonine-protein kinase B) mutation or amplification11,12. PI3Ks have three classes: class I, class II, and class III14,15; class I is most frequently linked to cancers15,16. Class I is a heterodimer and consists of two main subunits; the regulatory (p85a, p55a, p50a, p85b, p55c, or p101 isoforms) subunit and catalytic (p110a, p110b, p110d, or p110c isoforms) subunit16,17. Among the four catalytic isoforms, only phosphatidylinositol3-kinase, catalytic, α-polypeptide (PIK3CA) gene, which is found on chromosome 3q26.318, is frequently altered in cancer19,20.
In OC, PIK3CA gene mutation or amplification is present in >20% of women21. To the best of our knowledge, there are no published reports about PIK3CA gene amplification among Sudanese women with OC in Sudan. Therefore, the present study reports the amplification of the PIK3CA gene and its relationship with tumor type, grade, and ages in Sudanese women with OC.
This hospital-based, cross-sectional study included 90 adults patients (convenience sampling), out of which, 83 patients diagnosed with OC at Omdurman Maternity Hospital in the period 2013–2018. Another seven healthy ovarian tissues (taken from hospital records of cases diagnosed as unremarkable, which means no histological change) were used as controls to normalize the results. All cases were adults with no history for previous malignancies. The data was collected in the period between 2016 and 2018.
The Ministry of Health of Khartoum state, AlzaiemAlazhari University, and Omdurman Maternity Hospital Ethical Committees approved the study. Informed consent from patients was waived by the committees, since patients’ identity was anonymized, and only laboratory numbers were used.
Total RNA was extracted from the FFPE tissue sections using the total RNeasy FFPE kit from Qiagen© (Qiagen, Hilden, Germany), according to the manufacturer’s guidelines. To synthesize cDNA, QuantiTect Reverse Transcription Kit (Qiagen) was used according to the manufacturer’s information. The cDNA was stored at -20°C until use.
Amplification of the PIK3CA gene was assessed using quantitative real-time PCR (qPCR), performed in ViiA™ 7 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) using the QuantiTectSYBR Green PCR Kit (Qiagen). DNA content was standardized to that of long interspersed elements (LINE1), which is a repetitive element with similar copy number per haploid genome, both in normal DNA samples and cancer cells22. The primers for PIK3CA and LINE1 (reference gene as internal control) genes are shown in Table 1.
| PIK3CA | F, 5'-TATGGTTGTC TGTCAATCGGTGA-3' |
| R,5'-GCCTTTGCAGTGAATT-TGCAT-3' | |
| LINE1 | F, 5'CCGCTCAACTACATGGAAACTG-3' |
| R, 5'GCGTCCCAGAGATTCTGGTATG-3' |
The whole reaction volume of 20μl consisted of 10μl of SYBR Green Master Mix, 1μl forward and reverse primers (Macrogen, Korea), 1 μl cDNA, and RNA free water to complete the volume to 20μl. The following cycling conditions were used: 95°C for ten mins; 50 cycles with an annealing temperature of 55°C for 30s; and ending with an extension step at 72°C for 30s for both PIK3CA and LINE1 primers. A negative control (nuclease-free water as an alternative of cDNA) was incorporated in each run. A melting curve was obtained to provide evidence for a single reaction product. The relative quantification for PIK3CA gene was calculated using the 2- ΔΔCt method23. ΔΔCt represents the difference between the paired tissue samples (ΔCt tumor - ΔCt of normal tissue), with ΔCt being the Ct value for the target gene (PIK3CA) minus the Ct value for the reference gene (LINE1). Values > 2.0 were considered positive for gene amplification.
The results were analyzed using SPSS, version 24 (IBM SPSS). Descriptive analysis was performed for all variables (tumor type, grade, and age of women), which were taken from patient records. Fisher’s exact test was used between PIK3CA gene amplification and tumor types, grades, and ages for finding statistical significance. P-value < 0.05 was considered to be statistically significant.
Missing data in the dataset (Underlying data) concerning PIK3CA gene amplification were considered as undetectable values, which may be due to either a very small expression not detected due to the sensitivity of the method or completely absent, and therefore these samples were included in the analysis as no amplification.
This study included 90 ovarian samples. Among the 90 cases, the median age was 50 years (range, 18–75 years). All cases were distributed into two age groups: ≤ 50 years (range, 18–50 years), 48.2% (n = 40); > 50 years (range, 51–75 years), 38.6% (n = 32).
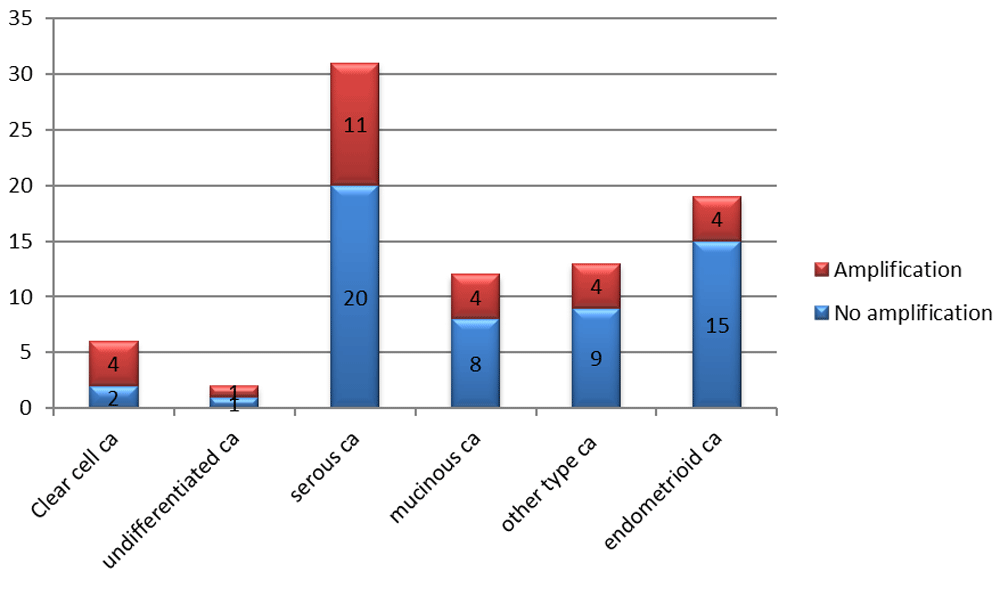
Amplification of the PIK3CA was common in 33.7% of OC samples (n = 28/83). The results of the amplification rate in different OC types is presented in Figure 1. A higher frequency of amplification was seen in high grade (39.5%; n = 17/43) rather than low grade (27.5%; n = 11/40) carcinoma (Figure 2). In addition, a higher frequency of amplification was seen in older (43.4%; n = 11/32) rather than younger (30.0%; n = 12/40) women. However, there was no significant association between carcinoma type or grade and age of women, and amplification of PIK3CA (Fisher’s Exact test p = 0.660, 0.698 and 0.687, respectively).
The phosphatidylinositol-3-kinase (PI3K) pathway plays a regulatory role in common physiological processes; when mutated, it contributes to cellular tumorigenesis, cancer evolution, and drug conflict14. In addition, it has a decisive role in the progression of OC2. Mechanisms for pathway activation are principally due to PIK3CA or AKT mutation or amplification11,12. Earlier studies from developed countries established that amplification was seen in the range of 13 to 24.5% of patients with OC24–28. A single study from a developed country with a small sample size (n=12) reported a higher frequency (40%)29. However, Abubaker et al. revealed a high-frequency level (35.5%) in individuals from the Middle Eastern with OC22, which supports the present study. This dissimilarity of the former study with the current study in the frequency may be due to the variation in sample size, ethnic background, the cancer biology in the Sudanese population or the sensitivity of the methods used.
Regarding the association of amplification with tumor grade; we observed an increased amplification frequency in higher grade carcinoma compared with low-grade carcinoma. Our observation implies that PIK3CA gene amplification is an early result in the progress of poorly differentiated (high grade) OC. However, amplification was not significantly associated with tumor type or grade in the present study. Our results are supportive of previous studies22,30 where no significant outcome was detected between histological tumor type or grade and PIK3CA amplification.
The small sample size limited our study. Further studies with an increased sample size and more additional data (e.g. stage at diagnosis, chemotherapy, radiation) are expected to verify the outcome of this study. The significance of combining other genes, which exist in the PIK/AKT pathway, should also be investigated.
The present study revealed that amplification of the PIK3CA gene occurs in about one-third of Sudanese women with OC, more frequently in high grade carcinoma and women of older ages. In support of recently published literature, our observations foster early reports that imply that oncogenic PIK3CA gene has a significant role in the progression and activation of OC and might offer a new strategy for specific targeted therapy and prognostic assessment in OC patients.
Figshare: pik3ca data.csv, https://doi.org/10.6084/m9.figshare.9121901.v531
Data are available under the terms of the Creative Commons Zero “No rights reserved” data waiver (CC0 1.0 Public domain dedication).
We are grateful to the staff at Omdurman Maternity Hospital (Department of Histopathology and Cytology) for their support. The authors express gratitude to the Institute of Endemic Diseases, University of Khartoum, Sudan, and Color-Lab (Qiagen), Egypt.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)