Keywords
Aging, Machine Learning, Age
This article is included in the Artificial Intelligence and Machine Learning gateway.
Aging, Machine Learning, Age
One of the fastest-growing areas at the intersection of clinical medicine and data science is the investigation of human aging1, with multiple avenues being explored to find biomarkers of aging that could be used to inform efforts to enhance human longevity2–4. If robust and easily-accessible biomarkers of aging are identified, they could assist in the rapid assessment of promising interventions aimed at increasing longevity, without the need to perform clinical trials that last decades. For instance, epigenetic modifications on DNA are increasingly being used to determine biological (rather than chronological) age, including how environmental determinants may affect an epigenetic signal for longevity4.
An individual’s biological age can be described based on the assumption that cellular aging processes, which are highly-influenced by the environment5, occur at different rates in different people with the same chronological age. As these ageing processes are associated with changes in routine biochemical measures6, algorithmic determination of biological or phenotypic age using widely-available indices such as those from blood test results is therefore becoming increasingly common. This has previously been done using both machine learning (ML) and statistical techniques3,6.
One important aspect for the utility of biological age measures is that a given output can be interpreted in order to guide individualized interventions. ML-based predictions of biological age have the potential to elucidate and describe complex, non-linear, and unintuitive patterns in biochemical data, which may provide greater predictive power compared to other statistical techniques. To date, published approaches to generate predicted biological age from biochemical data have used deep neural networks (DNNs), with the output being directly associated with mortality risk3. However, while individual outputs from DNNs are interpretable7, it is currently not possible to interrogate the effects of the entire training dataset on the model output, which may be important for determining how one may intervene given an individual’s output.
As a result of the issues with interpreting certain ML algorithms, the field of explainable artificial intelligence is developing rapidly8. If such approaches can be successfully applied to determining biological age from commonly available data, biological signatures of aging could be more rapidly discovered and tracked, including the ability to personalise interventions based on the outputs of the model. Here, we describe the development of an explainable ML model using blood marker data from the National Health and Nutrition Examination Survey (NHANES) database to predict biological age, as well as provide individual weighting for how each biomarker affected the final output. By determining how markers affect the model globally, potential target reference ranges associated with lower biological age can also be determined.
Data from a total of 46,739 participants (n=22,545 males and n=24,194 females) in the NHANES database were included, with a mean (range) age of 48.5 (19.0–85.0) years. A total of 39 common blood markers were used: complete blood count (CBC) with differential, lipids, fasting glucose, iron panel, and a comprehensive metabolic panel (including electrolytes, and liver and kidney function). Descriptive data for the dataset is listed in Table 1.
NHANES data (all available individuals with the 39 markers listed in Table 1 from years 1999–2015) was downloaded as .xpt files from the NHANES website using their in-built web search engine. The data was then concatenated, cross-tabulated, and stratified by gender. A random split in the data set was created to withhold 20% of participants (n=4,509 males and n=4,839) for model validation. The remaining 80% of the dataset was used to train an XGBRegressor model (XGBoost version 0.81) using chronological age and the 39 biochemical input markers. For the remaining 20% of the data, the 39 markers were provided to the algorithm9 with the chronological age withheld, and the resulting dependent variable “predicted age” defined as a measure of biological age. Age predictions for the withheld data were plotted against actual age using jointplot from the seaborn Python library (version 0.9.0).
For individual predictions, the weight of each marker was extracted using ELI5 (version 0.8.1), and graphed using a waterfall chart (version 3.8). For a given age prediction, each marker was individually weighted with regard to how it contributed to the final output. Shapley additive explanations plots (SHAP, version 0.26.0) were constructed to describe how each individual marker affects the predicted age output within the laboratory normal range.
To provide an individual output example based on data not seen by the algorithm9 previously, author C.K. had the necessary input markers measured by Quest Laboratories (Santa Cruz, CA). As C.K. is an author who ran his own data through the algorithm9 he trained during development of the manuscript, institutional ethical approval was not sought for publication of this data. C.K. approved the publication of his data in this manner.
Linear regression analysis (Figure 1) showed a significant correlation between predicted (biological) and actual (chronological) age (r=0.77 and 0.75 in females and males, respectively; p<0.0001 for both). However, discrepancies between the biological and chronological age could be considered clinically relevant, as they would allow for the generation of a signature of premature biological aging.
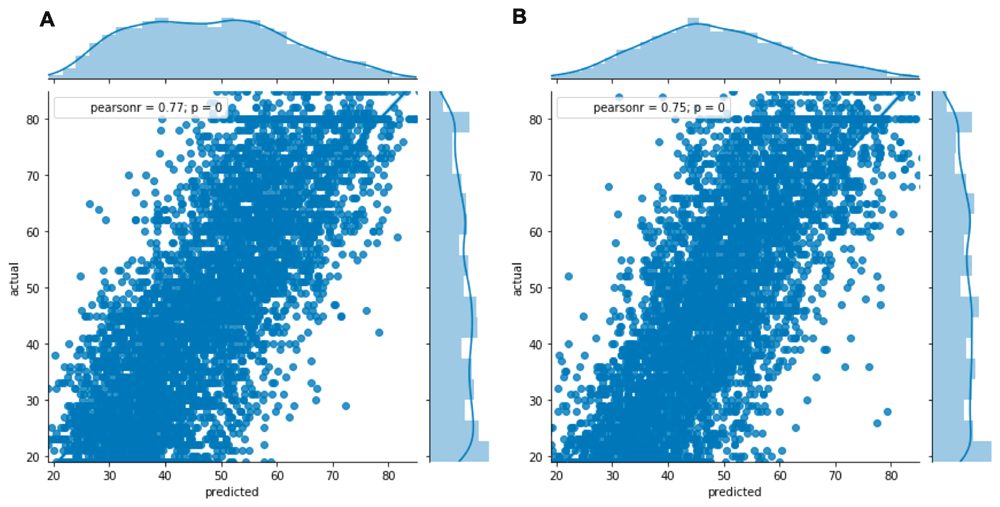
Data shown for women (A) and men (B) using the 20% withheld data (n=4,509 males and n=4,839). A significant correlation between predicted and actual age (r=0.77 and 0.75 in females and males, respectively) was seen in both sexes (p<0.0001).
SHAP summary plots (Figure 2) were used to determine which markers have the greatest influence on predicted biological age. The top 20 markers in terms of importance are shown. In females, blood urea nitrogen (BUN) had the greatest influence on biological age, with albumin the most influential marker in men. Fasting glucose was the second most influential marker in both sexes (Figure 2). SHAP plots for each of the 20 most influential markers are available on GitHub and Zenodo9. Based on each of these 20 markers, the level at which an inflection point was seen in the SHAP plot (i.e. when a further change in a marker would result in a net increase in predicted biological age) was determined, as well as the estimated range over which each marker would be associated with the lowest biological age (Table 2 and Table 3). Using the five most influential markers as an example, the lowest predicted age in women would be associated with a BUN 6–11 mg/dl, fasting glucose 71–86 mg/dl, bicarbonate (carbon dioxide) 19–22 mmol/l, total cholesterol 130–150 mg/dl, and mean corpuscular volume (MCV) 80–85 fl. In men, the lowest predicted age would be associated with albumin 4.6–4.8 g/dl, fasting glucose 70–88 mg/dl, BUN 6–12 mg/dl, red blood cell (RBC) 5.0–5.7 ×103/µl, and RBC distribution width (RDW) 11.0–12.5%.
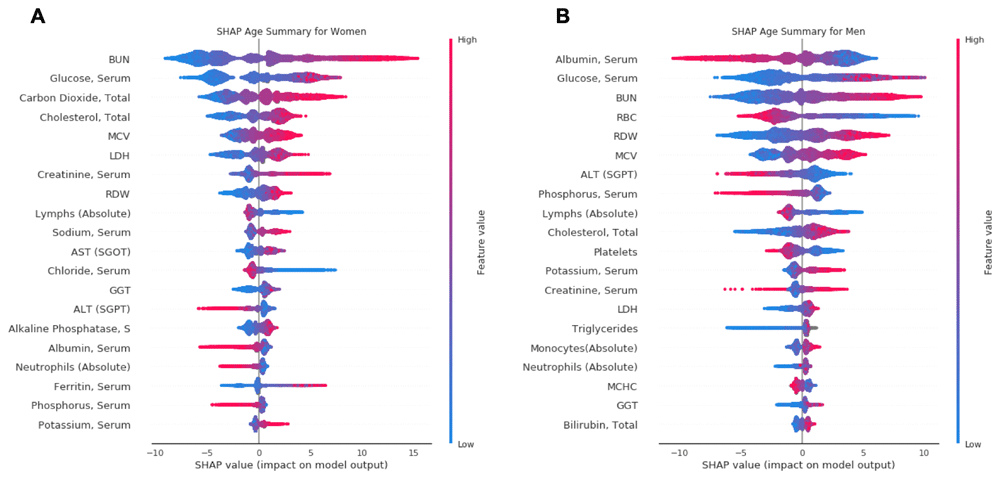
Data shown for women (A) and men (B). Each plot is made up of thousands of individual points from the training dataset such with a higher value being more red, and a lower value being more blue. This is depicted by the “feature value” bar on the right of each plot. Therefore, if the dots on one side of the central line are increasingly red or blue, that suggests that increasing values or decreasing values, respectively, move the predicated age in that direction. For instance, lower BUN values (blue dots) are associated with lower predicted age in both men and women.
Ranking of markers affecting predicted age in women, in order of importance, as determined by the SHAP summary outputs. Visual examination of the individual SHAP plots for each marker was used to estimate the range over which each marker would result in the lowest predicted age, and the magnitude of the adjustment in years. The final column is the value at which a marker changes from a net negative to net positive effect on biological age.
Ranking of markers affecting predicted age in women, in order of importance, as determined by the SHAP summary outputs. Visual examination of the individual SHAP plots for each marker was used to estimate the range over which each marker would result in the lowest predicted age, and the magnitude of the adjustment in years. The final column is the value at which a marker changes from a net negative to net positive effect on biological age.
For a given individual, the model output allows for each marker to be individually weighted with regard to how it contributed to the final output (Figure 3). The average age in the training dataset (BIAS) is given as a starting point, with each marker subsequently increasing or decreasing predicted age by a number of years. This allows for the most influential markers for the individual to be determined. The example shown is for one of the study authors (C.K.), the data for whom is available on Zenodo9. Bias (48.3 years) is sequentially adjusted, with the five markers contributing most to an increase in biological age were BUN (+3.5 years), total cholesterol (+2.8 years), potassium (+1.7 years), phosphorus (+1.2 years), and LDH (+0.9 years). The five markers contributing most to a decrease in biological age were lymphocytes (-1.2 years), RBCs (-2.3 years), albumin (-2.7 years) fasting glucose (-3.1 years), and triglycerides (-3.9 years). The final predicted biological age was 43.0 years.
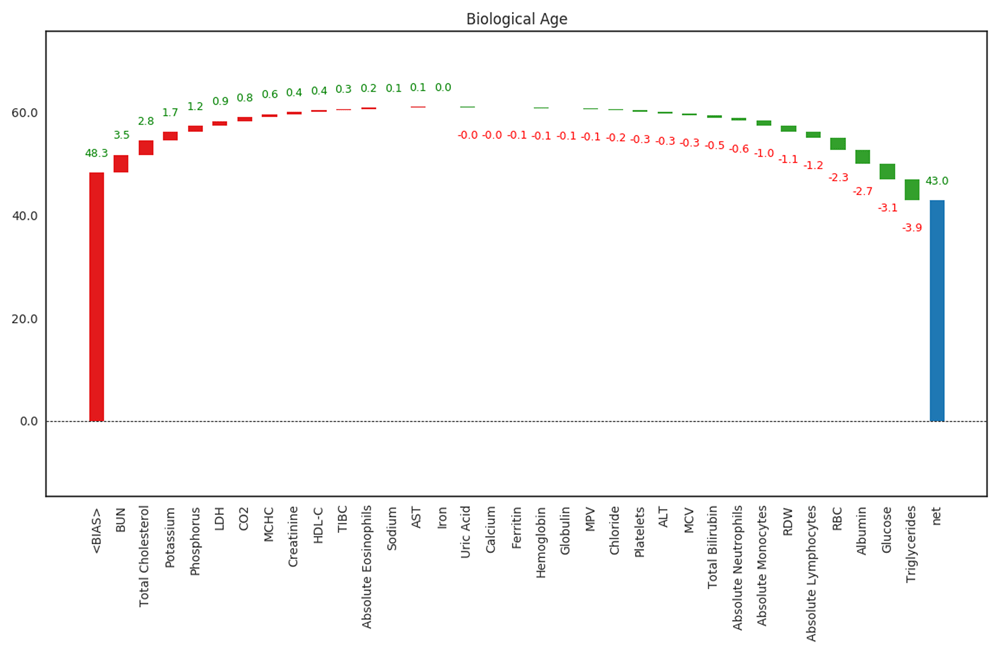
Bias (first column, 48.3 years) is the mean age in the input population. The five markers contributing most to an increase in biological age (columns 2–6 from the left) were BUN, total cholesterol, potassium, phosphorus, and LDH. The five markers contributing most to a decrease in biological age (columns 2–6 from the right) were lymphocytes, RBCs, albumin, glucose and triglycerides. The final predicted biological age (43.0 years) is in the last column.
Biomarkers of aging are increasingly important in the development and investigation of interventions with which to slow aging processes, which may also have the ability to aid in the treatment or prevention of aging-associated chronic disease. One such marker is the individual’s biological or phenotypic age, as reflected by patterns of biochemical markers in the blood, which have previously been shown to be associated with risk of mortality2,3,6. While there are a number of approaches to this problem in the published literature, we provide an alternative using a tree-based ML model that a) is fully interpretable, b) can be completely individualized for a given patient, and c) allows the development of target ranges associated with a potential signature for slowed biological aging.
One issue surrounding the utility of algorithmically-derived biological age is the response to any associated interventions or therapeutics. As this field is relatively new, it is uncertain how much an improvement in predicted biological age resulting from a given therapeutic approach will translate into improvements in longevity. Even if a given marker decreases predicted biological age, this also does not guarantee that manipulating the value will increase longevity. For instance, in our models, increasing ALT and decreasing total cholesterol were associated with lower predicted biological age; however, there are a number of scenarios where lower total cholesterol and higher ALT may be associated with increased mortality despite a lower predicted biological age10,11. Despite this, these models are at least able to generate hypotheses that can be tested in both the preclinical and clinical setting. Our approach also provides an example that other groups may use to produce fully-interpretable and personalisable outputs.
Though the current analysis does not include confirmation of the ability to predict mortality risk, certain outputs from the algorithm9 do provide some confidence that the output is likely to be associated with individual health outcomes. For instance, the greatest increase in predicted age associated with fasting glucose level occurs in the range 90–100 mg/dl, which is strikingly similar to the blood glucose level associated with the largest increase in mortality risk in multiple population studies12,13. Similar associations are seen with many of the target ranges derived from the algorithm9, such as for albumin, RDW, and ferritin (especially in men)14–16.
If modulation of certain markers does indeed contribute to the reversal of cellular aging processes, the combination of an individual output with the population SHAP plots for a given marker could therefore allow for targeted therapeutic interventions aimed at improving biological age based on an individual’s specific output. For instance, elevated fasting blood glucose could be decreased by addressing diet, exercise, micronutrient deficiencies, and reducing inflammation or psychosocial stress17. Similar approaches are also likely to improve cholesterol, RDW, and MCV, confirming that lifestyle factors should play a key role in the pursuit of health and longevity15,18,19. A personalised approach is important, because the markers contributing most strongly to biological age in the whole dataset are not necessarily the same markers that most strongly contribute to a prediction in a single individual (see example in Figure 3).
The current approach does have some limitations. The dataset may only be applicable in the United States, as different countries and ethnic backgrounds might display variations in both baseline biochemistry and predicted longevity3. Expanding available input data and allowing for stratification based on nationality and ethnic background will be the focus of future work. Larger and more expanded datasets will also allow for the analysis of biological aging in association with other potentially important factors such as genetics and the microbiota20,21. It is also worth mentioning that NHANES is designed to capture data that is representative of the US population. Therefore, this data comes from participants that represent a population that has some of the highest metabolic and cardiovascular disease prevalence in the Western world22,23, which may distort the results. Additionally, the current outputs would benefit from being correlated with disease outcomes or mortality in order to determine how well predicted biological age acts as an accurate biomarker of health and longevity.
By using well-understood and robust biomarkers that are available to almost any clinician, methods such as those described in this study can be used immediately as adjuncts to research investigating the outcomes of interventions designed to increase human longevity. As multiple methods are currently available with which to predict biological or phenotypic age, the field should also collaborate in an attempt to compare methods such that we can find the approach that results in an accurate output that can most easily be used in both the research and clinical settings.
All NHANES data used to produce the models is accessible through the CDC website (listed by NHANES study year): https://wwwn.cdc.gov/nchs/nhanes/search/default.aspx.
Data access, tabulation, and concatenation is automated by the “01-download-preprocess” Jupyter notebook file within our Zenodo repository; DOI: https://doi.org/10.5281/zenodo.24402039. This repository also includes the original Quest laboratory test results from author C.K., which were used to provide the worked example (Figure 3).
The algorithm developed here, including the associated libraries and the necessary versions, are available on Zenodo: https://doi.org/10.5281/zenodo.24402039.
License: GNU General Public License version 3
Notes: The algorithm itself can be trained and tested by running the “02-train-test-explain” Jupyter notebook. Note that each time the algorithm runs, a new random split in the dataset is generated in order to train and test the algorithm. Therefore, the resulting outputs might be slightly different.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
References
1. Levine M, Lu A, Quach A, Chen B, et al.: An epigenetic biomarker of aging for lifespan and healthspan. Aging. 2018; 10 (4): 573-591 Publisher Full TextCompeting Interests: PF is a founder and an employee of Gero LLC, the company is involved in development and commercialization of biomarkers of aging
Reviewer Expertise: aging research, biomarkers of aging, theory of aging, aging therapeutics
Is the work clearly and accurately presented and does it cite the current literature?
No
Is the study design appropriate and is the work technically sound?
No
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Partly
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
No
References
1. Zhavoronkov A, Mamoshina P, Vanhaelen Q, Scheibye-Knudsen M, et al.: Artificial intelligence for aging and longevity research: Recent advances and perspectives.Ageing Res Rev. 2019; 49: 49-66 PubMed Abstract | Publisher Full TextCompeting Interests: No competing interests were disclosed.
Reviewer Expertise: aging research, machine learning
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 04 Jan 19 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
https://www.ncbi.nlm.nih.gov/pubmed/27191382
So here it is the same study performed on a different dataset with just one method. And in that study, the comparison ... Continue reading Similar work was performed and published in 2016:
https://www.ncbi.nlm.nih.gov/pubmed/27191382
So here it is the same study performed on a different dataset with just one method. And in that study, the comparison with multiple other machine learning performed. Multiple reviews on this type of aging biomarkers were published since then, not sure why the authors chose to ignore them. Many other "aging clocks" were published since 2013 and there are common metrics for these clocks. For example, Mean Absolute Error (MAE). Since the work is not novel, it should at least provide a few case studies. For example, in cancer, BMT, etc.
https://www.ncbi.nlm.nih.gov/pubmed/27191382
So here it is the same study performed on a different dataset with just one method. And in that study, the comparison with multiple other machine learning performed. Multiple reviews on this type of aging biomarkers were published since then, not sure why the authors chose to ignore them. Many other "aging clocks" were published since 2013 and there are common metrics for these clocks. For example, Mean Absolute Error (MAE). Since the work is not novel, it should at least provide a few case studies. For example, in cancer, BMT, etc.