Keywords
Epithelial ovarian cancer, cross-sectional study, Immunohistochemistry, p53overexpression, Sudan
Epithelial ovarian cancer, cross-sectional study, Immunohistochemistry, p53overexpression, Sudan
Ovarian cancer is a fatal disease, the mortality rate ranks the highest of all gynecological malignancies1. It is considered the third most common cancer in the female reproductive system (following uterine cervix and corpus) and the leading cause of death from gynecologic malignancies in the United States as estimated by the American Cancer Society for the year 20191,2. Ovarian cancer refers to a group of morphologically and genetically heterogeneous neoplasms3,4. In Sudan, ovarian cancer represents the fourth most frequent tumor type in women5.
Based on clinicopathological and molecular studies, epithelial ovarian cancer (EOC) is classified as type I or type II. Type I tumors are genetically quite stable, typically present at a low stage, and reveal distinct, morphologic differences than type II tumors6. These include different histotypes: low-grade serous, endometrioid, clear-cell, and mucinous ovarian carcinoma. Type I tumors are characterized by distinct molecular genetics profiles, such as mutations in KRAS, BRAF, PIK3CA, PTEN and ERBB2, but not TP536. Type II tumors are generally high-grade serous (about 90% of all EOCs). They are highly aggressive, develop rapidly, present in an advanced stage in most cases, genetically unstable and express a mutated TP537–12. TP53 mutations have an important role in the prognosis and treatment of ovarian cancer13. Mutations in TP53 are found in high grade and rarely in low grade serous ovarian cancers. TP53 encodes the 53 kDa nuclear protein, their mutations leading to gain or loss of function of its protein product. TP53 mutation leads either to overexpression of p53 protein or complete lack of expression, while wild-type p53 is associated with focal expression14,15.
Immunohistochemical staining for p53 was considered as an essential biomarker for clinical trials targeting mutant p53 and used in the diagnostic workup of carcinomas of multiple sites, including ovarian cancers16. It is used as a substitute for TP53 mutational analysis, these mutations were global in high-grade serous ovarian cancer (HGSOC) (over 96% were mutated), so were used to discriminates between high- and low-grade serous carcinomas17–20. Access to TP53 sequencing is not feasible in many low- and middle-income countries; pathologists there used p53 immunohistochemistry, which is quick, easy to perform, inexpensive and can approach 100% specificity for the presence of TP53 mutation. Its high negative predictive value is clinically useful as it can exclude the possibility of a low-grade serous tumor18–20. To our knowledge, there are no published reports about the frequency of p53 immunostaining in type I and II EOC in Sudan. The study aimed to determine the frequency of p53 overexpression and its relationship with tumor types I and II and tumor grades among Sudanese women with EOC.
A cross-sectional, hospital-based study was implemented. All 114 available formalin-fixed paraffin-embedded tissue blocks (convenience sampling) previously diagnosed as epithelial ovarian cancer were collected during the period 2013–2016, in six governmental hospitals in Khartoum state, Sudan (The National Public Health Laboratory, Maternity Hospital, Military Omdurman Hospital, Alribat, Bahri, and Omdurman Teaching Hospital). Well-preserved tissue blocks with adequate tissue left for tissue microarray (TMA) procedure were included. Inadequate tissue blocks, and cases with missing tissue blocks were excluded. Slides from the original paraffin blocks were stained with hematoxylin and eosin (H&E), were reviewed according to 2014 WHO classification of ovarian tumors21, and were graded and typed according to the Kurman model8.
Available paraffin-embedded blocks from tumors were used for the construction of a tissue microarray (TMA). Representative areas of the tumor were identified and TMA blocks were constructed using two cores from each case. Sections were obtained from each TMA and were placed on negatively charged slides for immunohistochemistry.
Immunohistochemistry was performed to measure the protein expression of p53 monoclonal antibodies in ovarian carcinoma cases, as follow: Sections were cut into widths of 3–4 µm and placed on clean, electrostatically charged glass slides. Sections were dried by placing on a hot plate at 60°C for 15 minutes. Sections were dewaxed in two changes of xylene for two minutes. Sections were then hydrated through an ethanol series (100%, 90%, 70%, 50%) and water two minutes for each. Slides were retrieved using the water bath heat-retrieval technique22 and then treated with 3% hydrogen peroxide for 10 minutes. After that, sections were washed in phosphate buffer saline (PBS) (pH 7.4) for five minutes and treated with a 10% casein solution for 10 minutes. Sections were treated with ready-to-use primary antibody of mouse monoclonal antibody to p53 protein (clone DO7 IgG2b; catalog no AM239-5M; BioGenex, CA) for 30 minutes at room temperature in a humidity chamber, then rinsed in PBS before being treated with Super Sensitive polymer –HRP IHC Detection System (catalog no QD420-YIKE; BioGenex, CA) by incubated with enhancer reagents 15 min at room temperature, followed by a polymer-HRP reagent conjugated to anti-mouse and anti-rabbit secondary antibody for 15 minutes at room temperature, and rinsed in PBS. The entire antibody-enzyme complex is then made visible by incubation with a chromogenic substrate 3,3 diaminobenzidine for 7 minutes then washed in PBS for five minutes. For the staining step, sections were counter-stained in Mayer's hematoxylin for one minute washed and blued in running tap water before they were dehydrated through ascending concentrations of ethanol (50%, 70%, 90%, 100%). Sections were finally cleared in xylene and mounted using DPX. A known p53-positive breast cancer tumor was used as positive control. As the negative control, tumor specimens were immunostained under the same conditions without the primary antibody. Both the quantity of nuclear positivity and the staining intensity were measured in the immune slides examined under a light microscope (Olympus BX41, Japan). The intensity of staining was reported as negative, weak, moderate, or strong (0, 1+,2+,3+) in comparison with the positive controls (internal or external) and it indicated the average staining intensity of the tumor nuclei on the entire slide. An IHC score of p53 staining intensity was categorized as 0 for none, (no brownish color seen using x40 magnification), +1 for weak (brownish color seen using x20 and x40), +2 for moderate (brownish color seen using x10 magnification) and +3 for strong staining (brown color visible using x4 magnification).
The percentage of positive tumor cells was quantified by counting cells manually in at least 100 cells in 10 high power fields, averaged and categorized as ≥75% of cells considered as high overexpression, ≤75–50% considered as moderate expression and less than 50% considered as focal expression. Only 75–100% positive tumor cells with moderate and strong staining intensity considered as positive results.
According to the histopathological diagnosis, all 114 cases were EOC. They were classified as type I, type II and borderline, in 47.4% (n=54), 47.4% (n=54) and 5.3% (n=6) of cases, respectively. Grade of samples was classified as high in 53.5% of cases (n=61), low in 41.2% of cases (n=47) and non-graded tumors in 5.3% of cases, respectively (n=6). EOC histological subtypes were high-grade serous (HGS) in 54 cases (47.4%), low-grade serous (LGS) in six cases (5.3%), mucinous carcinoma (MC) in 22 cases (19.3%), endometroid carcinoma (EC) in 16 cases (14%), clear cell carcinoma (CCC) in seven cases (6.1%), malignant Brenner tumor in three cases (2.6%) and borderline in six cases (5.3%). Details of samples for each patient, alongside all unprocessed images, are available as Underlying data23.
Positive p53 immunostaining was seen in 35.1% (40/114) of ovarian epithelial carcinoma. From the 40 positive cases, staining intensity was as follows: 25 cases exhibited strong staining (+3), and 15 cases were moderate staining (+2). While the remaining 74 cases were considered as negative results (complete absence of p53 expression seen in 45 cases and +1 staining in 29 cases) (Figure 1–Figure 4).
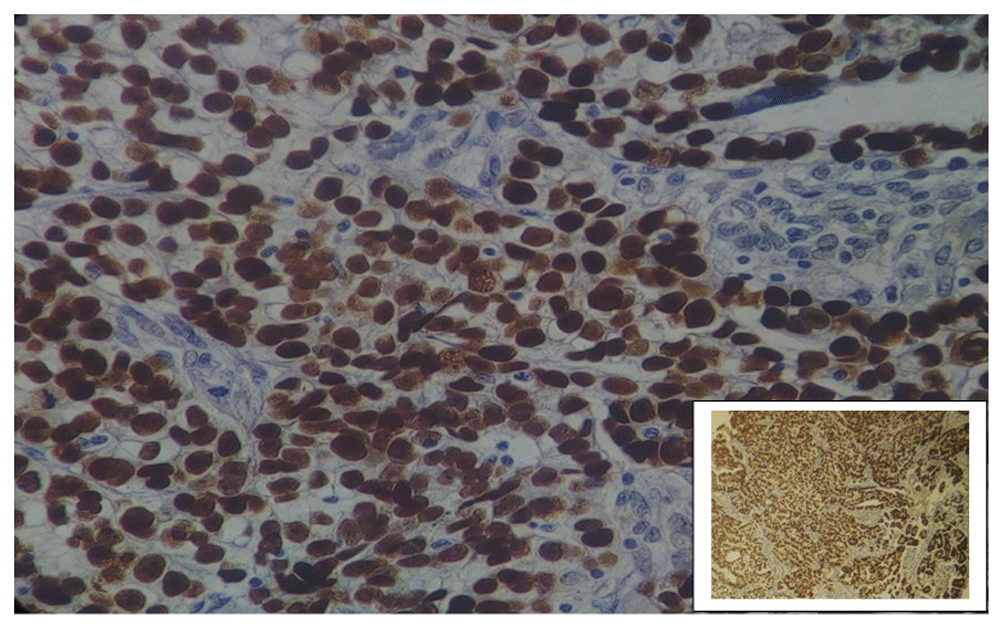
Magnification: main image, x40; inset, x10.
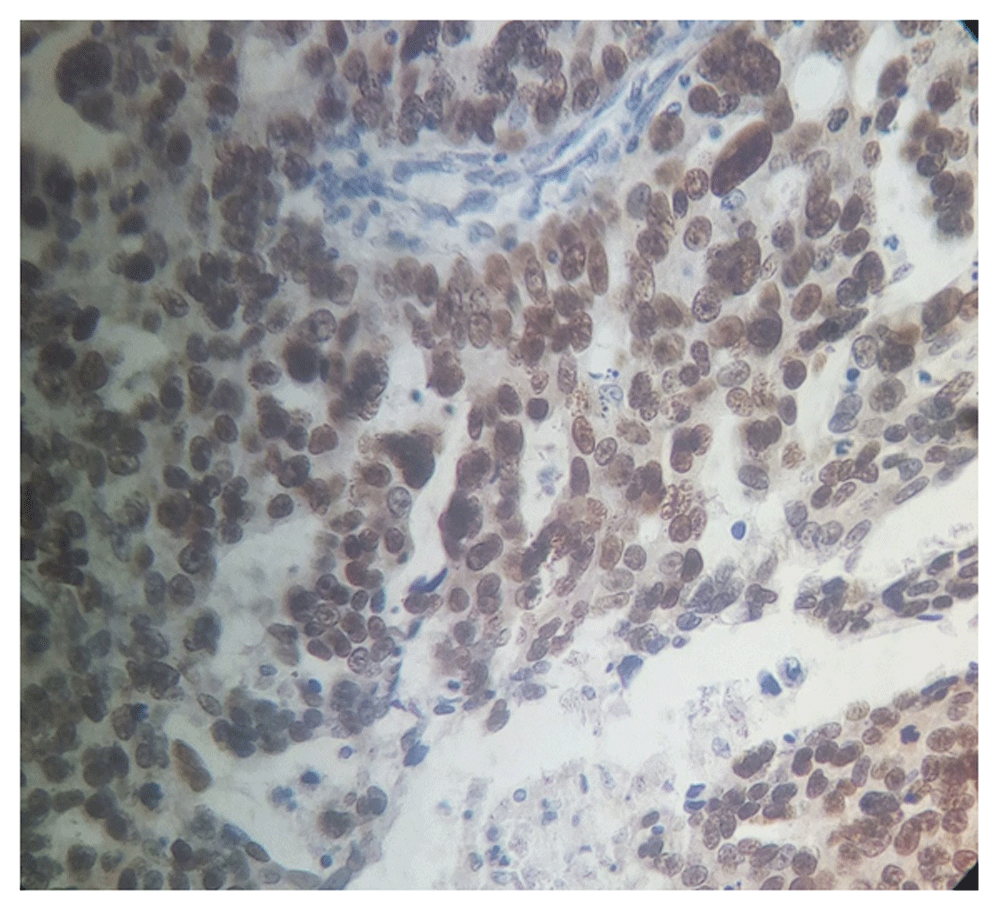
Magnification, x40).
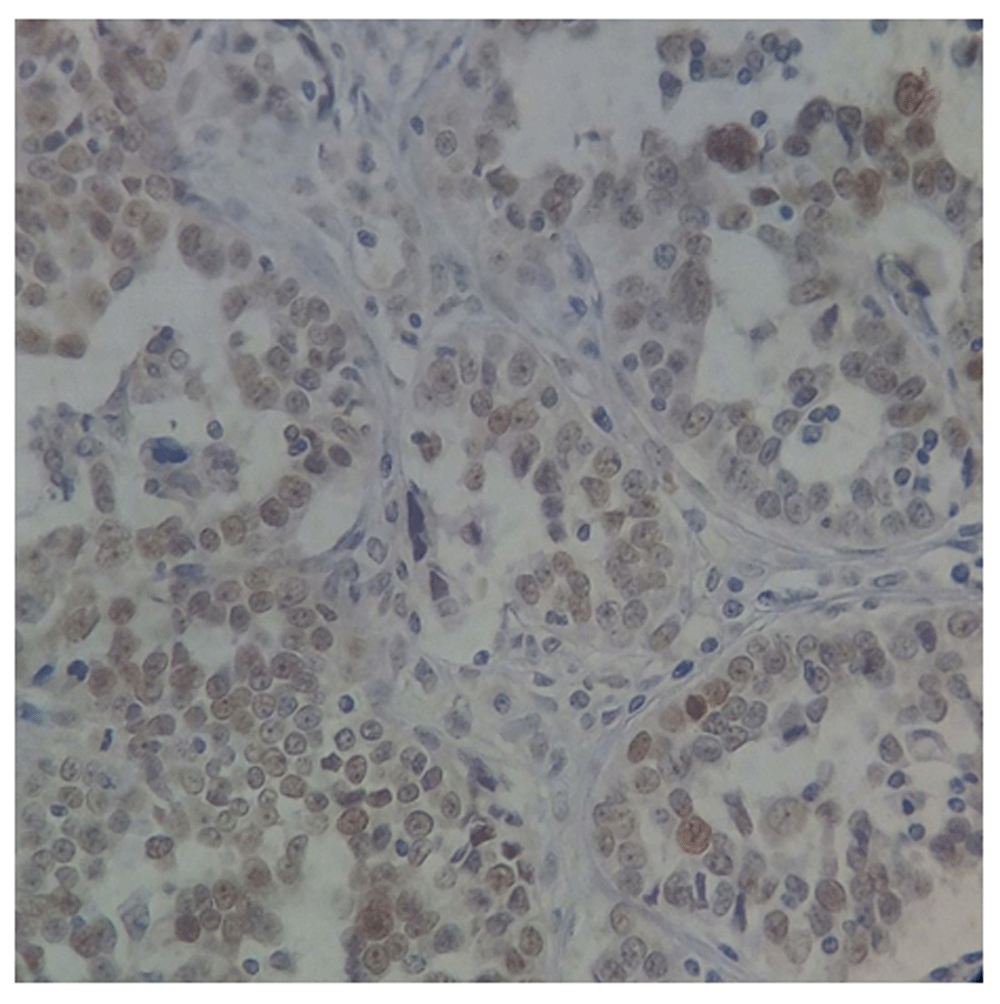
Magnification, x40.
Overexpression of p53 was associated significantly with the histological subtype (p = 0.004) as seen in Table 1. p53 expression was significantly associated with tumor grade (p = 0.003): it was positive in 49.2% (30/61) of high-grade tumors, 16.7% (1/6) of the non-graded borderline tumors, and 19.15% (9/47) of the low-grade tumors. The association between p53immunohistochemical expression and epithelial ovarian cancer type showed a highly significant association (p = 0.000). p53 was positive in 53.7% (29/54) of Type II and 18.5% (10/54) of Type I tumors (Table 2).
Immunohistochemical staining for p53 was used as a surrogate for TP53 mutational analysis to discriminate between high (over 96% was mutated) and low-grade serous carcinomas17–20.
In the present study, p53 positivity was observed in 35.1% (40/114) of EOC cases, and higher positivity was showed in HGS (53.7%; 29/54); a significant association was found between p53 expression and histopathological diagnosis and with tumor grade and tumor type. This result agreed with those of Tan et al., who found that 64.18% (43/67) of HGS samples analyzed overexpressed p5313. Harris et al., found that in 274 cases, 68% of tumors were characterized as p53 mutant (n= 186) and p53-mutant tumors were more likely to be HGS (72%) with significant association between p53 expression and histology and grade of tumor10. Markowska et al., found that positive expression of p53 protein was observed in 27.8% of EOC24. Yemelyanova et al. found out of the 57 tumors, 36 contained functional mutations and 23 cases (63%) with mutant Tp53 were positive for p53 IHC and 70% of HGS were p53 mutant16. Kobel et al. showed p53 marker overexpressed in 69% (118/171) of HGS cases18. Kobel et al. also showed that abnormal Tp53 expression was detected in 904 out of 912 (99.1%) of HGS25. Our results also agreed with those of Matsuo et al., who found that for 121 cases of high-grade serous p53 positivity was (71.4%)26, and Zhang et al., found that strong staining of p53 was observed in (70.8%) of serous carcinomas and showed that the tumor type was closely associated with the expression of p5327. Mozes et al., found p53 overexpression in 83.6% of HGS28. Bell et al. reported that Tp53 mutated in 96% of HGS29. Moreover, Oaknin et al. reported that expression of p53 markers in the various histological types of ovarian carcinoma (HGS 94%, LGS 0%, CCC 12%, EC 15% and MC 61%)17 were nearly to our results (HGS 53.7%, LGS 0%, CCC 14.3%, EC 12.5% and MC 31.8%). Lim et al., reported that p53 was not expressed in EC in 30 samples, but they agreed with our result in that they found HGS overexpressed p53 in 50% of cases examined30. Alexander et al. reported that p53 was positive for HGS in 68% of cases (52/76) and differed from our result in that they found positivity in LGS (24%; 18/76)19. Mackenzie et al., reported a higher percentage of p53 in mucinous carcinoma (68 %)31.
Furthermore, our result agreed also with Sallum et al., who reported that p53 positivity was found in 68.2% of HGS and significant association was found between p53 expression and tumor type15. Sundov et al.,32 and Tan et al.,13 also found that immunoexpression of p53 was significantly associated with tumor grade. And it disagreed with Brachova et al., who showed that p53 protein expression insignificantly associated with tumor grade33.
The present study showed that p53 marker was overexpressed in (53.7%) of type II, and (18.5%) of type I. This result agreed with Carter et al., 2018, who reported that p53 was highly expressed in type II EOC (68.8%) than type I (33.3%)4. HGSOC is the most frequent type of ovarian cancer and has been associated with a poor clinical outcome34. According to The Cancer Genome Atlas report, mutations in TP53 are the most common events in EOC, especially in HGSCs35.
The results of some of these studies may be conflicting primarily because of the indiscriminate grouping of TP53 mutations, which can result in either loss of function or gain of function36. GOF mutations can convert p53 protein from a tumor suppressor to an oncogene, leading to expression of a mutant p53 protein at a high level, while LOF mutants leading to loss of p53 protein expression37. TP53 mutations are classified according to their function as oncomorphic, loss of function and unclassified. Around 21% of all ovarian cancer patients harbor oncomorphic TP53 mutations, which had the highest p53 protein levels and contribute to chemoresistance and cancer progression, and the tumors with unclassified TP53 mutations express the mutated p53 protein at a fairly high level33. The differences found between the studies in the frequency of p53 expression may be due to the differences in scoring of p53 expression and interpretation of results: some studies scored this as overexpression (OE), complete absence (CA), cytoplasmic (CY) or normal/wild type (WT)18,20. Some authors consider complete absence of p53 expression as a mutant also because not all TP53 mutations alter the expression of the protein15,17,25. Complete absence of p53 expression does not indicate TP53 mutation, as a lack of immunoexpression may be found in normal cells16. So, we believe that true overexpression (more than 75% of the cells stained positive) was the most important type of mutation to consider due to their importance in clinical practice, as they are chemoresistant mutations33 and could also be interpreted easily in the immune-slide without any confusion.
The limitations of our study were related to the relatively small sample size. Many cases were found in the lab records but lacking tissue blocks, and some blocks contain less amount of tissue for TMA.
Our study showed that the overexpression of p53 tumor marker is associated with EOC, histological subtype and tumor grade, and found that high-grade serous tumors had a higher percentage of p53 expression in more than 50% of cases, while low grade serous was negative in 100% of the cases. We recommended the use of p53 immunohistochemical staining in the pathologic workup of ovarian carcinomas. Careful attention to laboratory protocols and practical works, including adequate controls, and training in interpretation is needed to make this a reliable test informing diagnosis and subsequent management of ovarian carcinoma.
Figshare: p53 data.csv, https://doi.org/10.6084/m9.figshare.990130723.
This project contains the following underlying data:
p53 analysis in ovarian cancer (spreadsheet containing details of analysis for each patient sample).
Uncropped, unprocessed images taken during this study.
Data are available under the terms of the Creative Commons Zero "No rights reserved" data waiver (CC0 1.0 Public domain dedication).
We acknowledge departments of histopathology and cytology at the six governmental hospitals for providing the work facilities. And gratefully thankful to the histopathology departments in the Universities of Alzaeim Alazhari and Sudan University of Science and Technology for hosting the practical work.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Partly
If applicable, is the statistical analysis and its interpretation appropriate?
Partly
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Molecular cancer research, cell plasticity
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Partly
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Statistics; Genetic Epidemiology
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 10 Oct 19 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)