Keywords
Depression, Alzheimer's, drug-induced depression, APOE, clinical decision support software
Depression, Alzheimer's, drug-induced depression, APOE, clinical decision support software
Alzheimer’s disease (AD) is the most common form of dementia1. It is a degenerative brain disease that eventually leads to death. Characteristic symptoms of the disease include difficulties with memory, language problems, agitation, and cognitive deficits that affect a person’s ability to perform everyday tasks. As the disease progresses, the burden increases and the individual will need help with even basic activities of daily living1.
An estimated 5.7 million people in the United States have AD, and over 35.6 million people are affected worldwide2. This number is projected to grow as the population aged 65 and older in the USA is anticipated to increase from its current level of 53 million to 88 million by 2050. Today, the total US spending on AD is estimated to be $277 billion1.
Research now suggests that the brain changes associated with AD begin as much as 20 years before initial symptoms3. Individuals often begin to show early signs of cognitive decline when they develop mild cognitive impairment (MCI). A systemic review of 32 studies found that 32% of people with MCI will go on to develop AD within five years1.
Like many chronic diseases, AD is not caused by one factor but instead develops because of multiple factors. Contributing factors include age, family history, the APOE ε4 gene4,5, as well as modifiable lifestyle factors1.
One factor often associated with AD is depression, both late-life (depression occurring after the age of 60 or 65) and recurring earlier-life cases. One in five individuals experience at least one depressive episode in their lifespan6, and it occurs more often in older populations2,7. In 2009–2012, depression was estimated to affect more than 5% of all US adults8. Depression has also been found to be more common among women9. Many people with depression go undiagnosed and untreated8.
Evidence suggests that depression is preventable and treatable, and may be a modifiable risk factor for preventing or delaying dementia in later life7. Depression can lead to cognitive deficits in some individuals, sometimes referred to as “pseudodementia”2,10. Older adults are at an even greater risk of developing pseudodementia9,11.
It is currently unknown if depression is a risk factor for developing AD, or a prodromal sign of the disease2,5,9,12–14. The weight of the evidence suggests that depression is likely a risk factor, though2,10,15. This question remains partly due to the timing of studies that have been conducted in late-life, the variability in follow-up time, and also the fact that depression frequency and duration times are often not recorded2.
Research has established that depression is associated with a greater risk for AD. Many studies have concluded that depression doubles an individual’s risk of developing dementia, particularly AD7,9,10,16,17. Other studies on depression and AD have shown that those who develop depression are 1.5 times more likely to develop AD compared to those who are not depressed9. The evidence is not yet conclusive as to if early or late-onset depression might be a greater risk factor, but the severity of late-life depression has been found to be an important risk factor for dementia2,7. Some studies have shown that depression in earlier life is more closely associated with vascular dementia, and depression in later life is a greater risk for developing AD18.
APOE ε4 status has been linked to higher rates of depression and is a definitive risk factor for AD5,19. A study completed in India found that those with APOE ε4 had a 4.7 times higher risk of developing depression in old age20. Populations in Sweden with the APOE ε4 gene also had a higher risk of developing depression. Furthermore, APOE ε4 is associated with more severe depressive symptoms4. Post-mortem studies have reported greater hippocampal amyloid plaque and neurofibrillary tangle pathology in AD patients with a history of depression than those without21.
While the specific pathway, or pathways, linking depression and AD have not been defined yet, many mechanisms have been hypothesized. Depression may lead to an increase in amyloid plaques through the depression-associated stress response. This response ultimately results in an increase in β amyloid production22. Chronic inflammation also plays a significant role in the pathophysiology of AD23 and is seen in both depression and AD24. It is hypothesized that stress leads to chronic inflammation in depressed persons. Neuroendocrine changes comparable to those found in chronic stress have also been found in depression and AD models10. Additionally, pro-inflammatory cytokines have been shown to interfere with serotonin metabolism, leading to a reduction in synaptic plasticity and hippocampal neurogenesis22,23. Increased cortisol production can also lead to atrophy in the hippocampus and cognitive deficit25. Interleukin-6 (IL-6), C-reactive protein, and tumor necrosis factor are all increased in depression and may be risk factors for dementia26. Brain-derived neurotrophic factor (BDNF), often associated with synaptic plasticity, is decreased in depression and AD27. Furthermore, MRI studies have found that hippocampal atrophy occurs in cases of depression, suggestive of AD pathology28. Lastly, many lifestyle factors associated with depression are also known risk factors for AD, including poor diet, lack of physical activity, and low social engagement1,9.
Of the studies conducted on depression and AD, this author could not find one that examined the link between depression, AD, APOE, and depression-inducing-drugs (DIDs). Drug-induced depression, also referred to as substance-induced mood disorder in the literature, is a well-known side effect of many medications. The first notion of a “depressogenic” drug was documented in 1954, referring to long periods of treatment with the antihypertensive drug reserpine29.
Older populations have a greater risk of exposure to DIDs, as they are more likely to be on multiple medications8,30–32. It has been found that the use of multiple medications increases the risk of simultaneous depression and that older age is significantly associated with the use of DIDs8.
The most common DIDs include narcotics, benzodiazepines, anticholinergic medications, antidepressants, sedatives, beta-blockers, calcium channel blockers, and oral contraceptives8,30,33. The mechanisms for a few depressogenic drugs have been studied, and their additive risk factor for AD cannot be ruled out. In one longitudinal study, seven out of 15 individuals who developed AD had been taking antidepressants5.
Possible mechanisms for DIDs have been proposed34. These mechanisms include inhibiting calcium-dependent neurotransmitter release by blocking the inflow of calcium into cells35, decreased serotonin36, reduction in G-protein-coupled catecholaminergic or cholinergic receptors37, decreased levels of dopamine38, and displacing nicotine from acetylcholine receptors38. Because of the wide range of drugs that can cause this side effect, there is “no known reason to presume that all drugs produce depression via a common pathophysiological mechanism”34.
Drug-induced depression is more likely to occur in those who have a higher risk factor for depressive disorder, as may be the case for individuals with the APOE ε4 allele. For those already predisposed to depression, such as carriers of APOE ε4, DIDs might have an amplified effect, leading to more frequent occurrences of depression.
Here, the author examined the prevalence of DIDs in elderly non-symptomatic and symptomatic AD patients to find how many DIDs the population was taking, the prevalence of depression, and whether APOE ε4 status has any effect on depression levels with concurrent DIDs.
The data presented here is from uMETHOD Health’s (uMH) database of patients. uMH has created a precision-medicine platform to create personalized treatment plans for patients with and at risk of dementia and AD39. All individuals self-selected to follow their treatment plan. The treatment program was recommended to them by their physician, or they found uMH online and chose to purchase the personalized evaluation and treatment.
uMH’s platform identifies and addresses active issues and creates repeatable and practical treatment plans for use in doctors’ practices.
The bioinformatics platform’s internal software is written in the Python language. It interfaces to an external portal, used by the medical and coaching teams to gather input, and returns reports written in PHP and Java. External medical databases support internal rules-processing algorithms. These are sourced from bodies such as the National Institutes of Health (NIH), Food and Drug Administration (FDA), pharmaceutical trade groups, and consortia focused on topics such as genetics or allergies.
Natural language processing (NLP) techniques are used throughout the input steps, particularly for precise identification of lab tests, medications, drug indications, and comorbidities. A range of NLP techniques are employed to normalize input data. The algorithms that implement this information platform go through a consistent set of steps each time they load a person’s input to generate a new set of reports. These steps are rules-based, so the logic and evidence sources can be tracked (and evaluated).
The platform was used as a source of data only and was not used for data analysis in this study.
The data set consists of all the patients in uMH’s database who were 65 or older.
The inputs to uMH’s data-analytics engine comprise genomics, bio-specimen measurements, medical histories, medications, allergies, comorbidities, lifestyle data, and cognitive evaluations. Data was collected from physicians, patients, and caregivers from annual wellness visits and through uMH’s patient forms. Forms were completed online via uMH’s secure portal or in paper format, depending on the individual’s computer skills and access. Physicians were instructed to follow a set of inclusion and exclusion criteria from Keine et al. 201839 before enrolling patients.
Each patient was evaluated by a physician to determine if they were symptomatic for AD following standard of care guidelines40. This evaluation included administering a baseline cognitive test to determine patients’ cognitive status at the time of enrollment. The physician chose to administer either the Self-Administered Gerocognitive Exam (SAGE) or the Montreal Cognitive Assessment. Cognitive impairment was determined following each tests’ specific guidelines41,42. Tests were scored at the physician’s office by trained personnel, and completed tests were either uploaded to uMH’s secure portal or were sent to uMH by the physician’s office to be entered into the database. The physician then used the findings to qualify the patient as symptomatic or non-symptomatic in uMH’s database.
Genomic data was obtained through consumer-focused companies such as 23andMe or Ancestry.com. Interested patients purchased a genomic kit through the company of their choice and followed the procedures outlined by the genomic testing company. Raw datasets were accessed by the patient directly from the genomic company’s website and uploaded to uMH’s secure portal. As this was an optional step, many people chose to complete their analysis without genomic information. Therefore, APOE ε4 status is only known for a subset of the population presented here.
Depressive symptoms were determined through patients’ self-reporting, the Short Form Geriatric Depression Scale (SF-GDS) at the physician’s office43, prior physician diagnosis as indicated by the patient’s medical history, or a prescription for antidepressant medication.
DIDs were identified from Qato et al. Table 28. The DIDs listed here were coded into uMH’s bioinformatics platform so that each patients’ medication list from uMH’s database was checked against this published list by the platform’s algorithms.
The data was analyzed using statistical methods to examine the prevalence of DIDs and depression in an elderly population currently at risk of or diagnosed with AD. The frequency of DIDs, antidepressant medications, and depression was analyzed against gender, cognitive impairment levels, depression symptoms, and current medications. This information was then subsequently compared to APOE status to investigate if a possible relationship exists between the APOE ε4 allele and depression levels, with or without DIDs.
All statistical analyses were performed using SAS University Edition. Power calculations for independent, two-sided t-tests were completed to compare population means. Either pooled or Satterthwaite P values were used based on equality of variance testing. Chi-square tests were completed for categorical data. Means, standard deviations, and frequencies were established using SAS. Significance was determined using a 95% confidence interval.
Following the guidelines put forth in the 21st Century Cures Act44, Clinical Decision Support Software (CDSS) is not regulated by the FDA, and uMH’s treatment protocol does not include any investigational drugs. Because of this, the platform was taken straight to market and did not need approval by an ethics committee or board.
uMH’s algorithm platform automatically assigns every participant a random eight-character ID when they are first entered into the system to de-identify their information. All data was collected under their randomly assigned ID, and the investigators here only had access to this de-identified information.
The data presented here is an analysis of data already gathered from patients; no experiments were performed on humans. Each participant self-selected to participate under the guidance of their individual physician. No patients were recruited by uMETHOD Health. All patients gave consent upon initial data collection and were informed that their de-identified information would be used for future research purposes. For patients who presented with cognitive decline, their caregiver or power of attorney was also informed of all data collection and gave consent to use de-identified data for future research.
Utilizing uMH’s data set, 292 individuals were analyzed for this paper (Table 1). A total of 189 people chose to provide genomic data. Of these 189, 111 have at least one copy of the APOE ε4 allele (Table 1). Those with the APOE ε4 allele are referred to as ε4 and those without are referred to as nε4. This population took an average of 11.51 (SD 8.86) medications per person, and 1.16 (SD 1.27) DIDs per person. The maximum number of DIDs seen for one person was five (Table 1).
36.21% of the population had depressive symptoms (Table 1). Individuals with depression were significantly more likely to be taking at least one DID (71.15% vs 28.85%, P value 0.005) (Table 2 and Figure 1). Those with ε4 had a significantly higher rate of depression than nε4s (69.12% vs 30.88%, P value 0.03) (Table 2). Females were also more likely to be depressed than males (65.71% vs 34.29%, P value 0.005) (Figure 2). The same held true when APOE ε4 status was considered with gender (68.09% vs 31.91%, P value 0.002) (Table 2).
60.56% of the total population was taking at least one DID (Table 1). Females were more likely to be taking a greater number of concurrent DIDs than males (1.29 [SD 1.32] vs 0.99 [SD 1.19], P value 0.045) (Table 2 and Table 3 and Figure 2). Those taking a DID were more likely to be taking a higher number of antidepressants (0.58 [SD 0.88] vs 0.2 [SD 0.46], P value <0.0001) (Table 2 and Table 3).
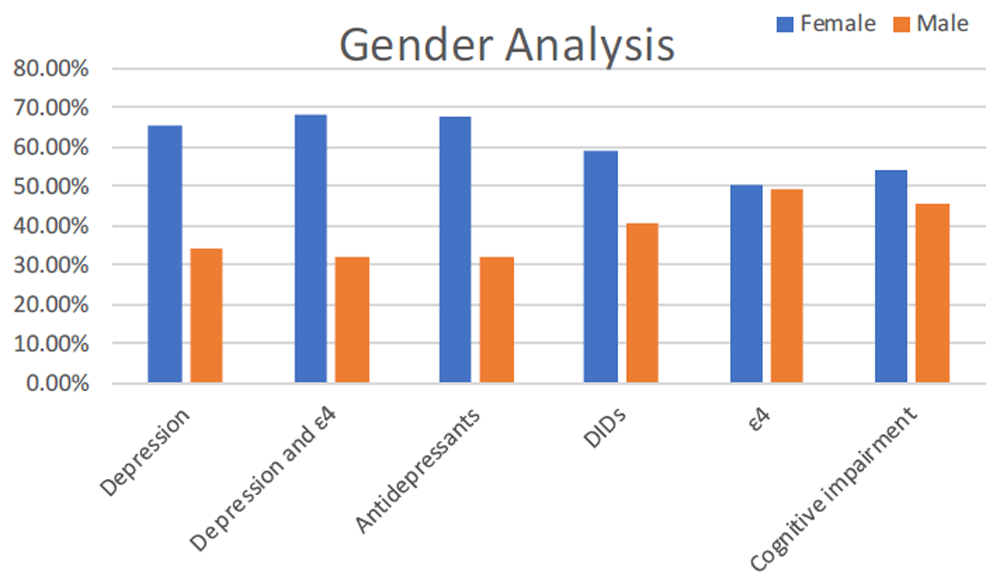
Analysis of the percentage of each gender with depression, depression and APOE ε4, taking antidepressant medication, taking a depression-inducing drug (DID), possessing the APOE ε4 allele, and measurable cognitive impairment.
29.58% of the overall population was taking at least one antidepressant medication (Table 1). Those taking an antidepressant were more likely to be taking concurrent DIDs (77.38% vs 22.62%, P value 0.0002) (Table 2). Gender was also a significant factor for antidepressants. Females were more likely to be on at least one antidepressant (67.86% vs 32.14%, P value 0.005) (Figure 2), as well as taking a greater number of concurrent antidepressants (0.52 [SD 0.80] vs 0.32 [SD 0.71], P value 0.029) (Table 2 and Table 3).
A higher, though not statistically significant, number of people taking antidepressants were more likely to have ε4 (70% vs 30%, P value 0.084) (Table 2).
75.33% of this population had measurable cognitive impairment (Table 1). Those who were impaired took a higher number of concurrent antidepressant medications (0.46 [SD 0.83] vs 0.25 [SD 0.52], P value 0.036) (Table 2 and Table 3). There was also a higher percentage of cognitively impaired individuals taking antidepressants, but the number did not reach significance (80% vs 20%, P value 0.316). A higher percentage of people with cognitive impairment were also taking DIDs (57.49% vs 42.51%, P value 0.394), but again the number did not reach significance (Table 2).
DIDs are a substantial clinical, medical, and public health problem34 that has yet to be addressed in older populations. DID consideration is especially important in populations with an increased risk or a current diagnosis of AD. Prescriptions for elderly patients, especially those with a family history of AD or who currently have symptoms of cognitive decline, should be closely monitored and should avoid taking DIDs whenever possible.
The results of this work support that ε4 populations are at a greater risk of depression. Whether depression is a prodromal symptom or a risk factor for AD cannot be concluded from this data, but many other studies provide compelling evidence that depression may contribute to a person’s risk of developing AD. If depression is a precursor to AD, then DIDs may accelerate the clinical presentation of AD. It is worth further study to see if those who possess the ε4 allele should avoid drugs likely to cause depression symptoms, as they may be predisposed to develop depression. Further research into the possible mechanisms of DIDs is also warranted and could possibly help further elucidate the correlation between depression and the development of AD.
This work also showed that DIDs are abundant in elderly individuals’ medication regimes. Re-evaluation of medications that have been documented to cause depression could potentially lead to better cognitive function for many patients. This may be especially beneficial to the ε4 population. Further research may improve guidelines for prescribing DIDs to elderly patients, and additional specifications may be needed for those with the ε4 allele.
Females appear to be at an even greater risk for DIDs as this study found they may already be pre-disposed to depression, with the risk increasing with the ε4 gene, and are more likely to be taking DIDs and taking a higher number.
Further research is warranted to see if patients with different levels of cognitive impairment respond differently to DIDs, or if there are other contributors to developing depression as a side effect. Other factors such as age, body mass index, and comorbidities could also be explored to see if they impact patients’ risk of developing DID.
With the rising numbers of elderly in the US, along with concurrent use of multiple prescription drugs, DID burden on cognitive health continues to grow. 30% of older adults in the United States take five or more medications daily45, known as polypharmacy. Each additional drug increases a patient’s risk of experiencing adverse effects, including depression and cognitive impairment.
CDSS provides a reliable and reproducible method to help with polypharmacy and DIDs. CDSS is software that is created to provide decision support for the diagnosis, treatment, prevention, cure, or mitigation of diseases or other conditions44. The goal of CDSS is to provide clinicians with actionable recommendations to improve clinical decision making46,47. One considerable advantage of CDSS is its inherent ability to process immense amounts of data quickly. This makes CDSS ideal for dealing with complicated medication regimes.
Machine-learning algorithms, one element of CDSS, can be equipped to help physicians and patients better curate their medications to prevent adverse events, unwanted side effects, and even duplicated prescriptions. It’s estimated that 25% of people aged 65–69 take at least five prescriptions, and it is not uncommon for people to take 20 or more drugs48. One individual in this dataset was on a total of 67 medications. The considerable number of patients with polypharmacy and an average of 32 new drugs approved by the FDA each year make this an insurmountable task for a physician to take on by themselves49.
CDSS can quickly review a patient’s medication list, no matter how large, matching it to current guidelines to provide physicians with reproducible guidance. This guidance could include recommendations for deprescribing medications that are therapeutically repetitive, medications that interact with other drugs, and medications that interact or have diminished therapeutic benefit due to an individual’s genetics. CDSS can also alert physicians to medications such as DIDs and anticholinergic drugs that have been shown to cause cognitive burden for elderly patients. CDSS also has the ability to suggest formulary alternatives to reduce the amount of DIDs in a medication list.
CDSS has the potential to support pharmacogenomics. Pharmacogenomics is the study of identifying genomic variants that affect drug efficacy and pharmacokinetics (PK) or pharmacodynamics (PD)50. There are many examples of medications that are effective or ineffective for patients based on their genome, including antidepressants. As of 2015, 20 genetic variations have been found that can affect around 80 medications50. In fact, one gene can affect multiple medications. Many of uMH’s clients choose to get their genome sequenced to find out more about their genetic risk and protection against AD. This same data can also be used to help select the most beneficial medications for each person.
uMH’s bioinformatics platform currently mines publicly available databases to check for drug-gene interactions (DGIs). The platform analyzes more than 2,000 single nucleotide polymorphism (SNPs) per individual, some of which pertain to DGIs, others are used to quantify the genetic risk of AD as well as other genetic diseases.
DGI information is drawn from three sources of pharmacogenomic information: the Clinical Pharmacogenetics Implementation Consortium (CPIC) effort51, DrugBank52,53, and SNPedia’s SNP and genoset compilations54,55. DGIs are looked at in two categories: genetics that influence how enzymes metabolize each drug, and relationships between specific genes and drugs. Each medication a person is currently taking is compared against a table of genes that affect it, and these genes are, in turn, compared to those in the person’s genome.
Approximately half of the people placed on an antidepressant medication do not show signs of improvement and over half will experience an adverse side effect. This is partly due to each person’s genetic makeup and its impact on the PK and PD of the medication56. Many pharmacogenomic studies have been conducted on antidepressant medications and found that their efficacy is highly correlated with SNP variations. Metabolizing enzymes such as P450 and the CYP2D6 polymorphism have both been found to have an effect on the drug plasma levels of antidepressant medications57.
Precision-medicine platforms, like uMH’s CDSS, offer the opportunity to improve the fitting of a drug to a patient with less trial and error, based on their genetics. This would allow for patients to be placed on the most effective medication as soon as possible, potentially speeding up recovery time from depressive symptoms.
CDSS could also be made more effective if linked to systems pharmacology models. These models have been developed to help predict in vivo drug effects in biological networks58,59. This approach seeks to look at the whole, instead of the more traditional single transduction pathway of drug administration and response. Drugs often have off-target or secondary effects that can be hard to predict as multiple systems can be involved and must be accounted for. Patients’ responses to drugs are also inherently variable due to their unique genome, environment, disease state, and the high prevalence of polypharmacy32,58.
A systems biology analysis could be useful in further defining the mechanisms behind DIDs. This information could then further inform future research into potentially lessening the depressive effects of these drugs, and also inform research into whether depression is a true risk factor for AD or a prodromal symptom.
Many older adults who experience late-life depression never fully recover from their depression-induced cognitive impairment, even after successful treatment of the depression. All these observations magnify the urgent need to provide better vigilance when prescribing medications for the elderly.
A more technology-friendly view is needed when it comes to managing complex medication regimes, especially when medications that can affect patients’ cognitive abilities are involved (as evidenced by the lack of publications on DIDs and the AD population).
The findings presented here support numerous previous publications indicating a relationship between depression and cognitive decline. While our observational data can only provide conjecture and find correlations, it is clearly worth closely monitoring DIDs in elderly populations. It is of even more importance when a patient may already be at increased risk of AD. There is a better path forward for patients with precision medicine and CDSS.
The data that support the findings of this study are available from uMETHOD Health but restrictions apply to the availability of these data, which are proprietary company information, and so are not publicly available. Data are, however, available with permission of uMETHOD Health. Requests will be granted on a case-by-case basis to researchers affiliated with an accredited institution following the submission of an ethically approved and relevant research proposal. Readers and reviewers can request access to the dataset used in this manuscript by sending a request to info@umethod.com. The National Alzheimer’s Coordinating Center (NACC) database is a publicly available dataset that is representative of the dataset analyzed in this study and can be used to apply the methodology described in this manuscript.
Authors had full access to all of the data in this study and take complete responsibility for the integrity of the data and accuracy of the data analysis.
Thanks to Dr. John Q Walker and Mark Zelek for their support.
Dr. John R. Cirrito confirms that the author has an appropriate level of expertise to conduct this research and confirms that the submission is of an acceptable scientific standard. Dr. John R. Cirrito declares they have no competing interests. Affiliation: Department of Neurology, Washington University, St. Louis, USA.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
I cannot comment. A qualified statistician is required.
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Psychopharmacology
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
No
If applicable, is the statistical analysis and its interpretation appropriate?
Partly
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: CGS is an employee of Predemtec AG.
Reviewer Expertise: biochemistry, molecular neuroscience, biomarkers, Alzheimer´s disease, diagnosis of Alzheimer´s disease
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 22 Oct 19 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)