Keywords
Uropathogenic E.coli, Phylogenetic groups, Clermont’s, Quadruplex PCR, Antimicrobial resistance.
This article is included in the Pathogens gateway.
This article is included in the Antimicrobial Resistance collection.
Uropathogenic E.coli, Phylogenetic groups, Clermont’s, Quadruplex PCR, Antimicrobial resistance.
Urinary tract infections (UTIs) remain a major cause of morbidity, with over 1.5 million annual cases reported worldwide (Lee & Neild, 2007)(Neild, 2003). Escherichia coli is the major cause of UTIs accounting for over 80–90% and 30–50% of all community-acquired and hospital-acquired UTIs respectively(Foxman, 2010). Phylogenetic (evolutionary) groups of E. coli strains have been shown to differentiate between pathogenic and commensal strains depending on their fitness landscapes and virulence characteristics. Although multi-locus sequence typing (MLST) and ribotyping are the gold standard methods for phylogenetic typing of E.coli strains, these methods are expensive, time-consuming and require the collection of typed strains (Clermont et al., 2000).
In 2000, Clermont et al (Clermont et al., 2000). developed a triplex PCR assay that classified E.coli strains into four different phylogenetic groups, A, B1, B2 and D based on the presence or absence of two genes, namely chuA, yjaA, and one DNA fragment TspE4.C2. Since 2000, growing knowledge of MLST for E.coli from different habitats has made it possible to validate the triplex PCR method (Gordon et al., 2008). The validation studies found that only 80–85% of all E. coli phylogenetic groups were assigned correctly(Gordon et al., 2008)(Clermont et al., 2013).
In 2013 Clermont et al. (Clermont et al., 2013) added an additional gene target, arpA, to the three candidate markers (chuA, yjaA and TspE4.C2) and developed a quadruplex PCR assay to classify E. coli isolates into eight phylogroups: A, B1, B2, C, D, E, F, and clade I/II. The use of this quadruplex PCR phylotyping method has been found to correctly assign 95% of all E. coli strains. Phylogenetic analysis has demonstrated a relationship between different E. coli phylogenetic groups, antimicrobial resistance and other virulence characteristics (Iranpour et al., 2015). Different studies have shown that the most virulent and antimicrobial-resistant extra-intestinal E. coli strains belong mainly to group B2 and, to a lesser extent, to group D (Iranpour et al., 2015)(Bashir et al., 2011)(Liu et al., 2014). In contrast, most of the commensal strains are associated with group A or group B1 (Clermont et al., 2000). In this study, we aimed to determine the prevalence of the different phylogenetic groups of Uropathogenic E. coli strains using the new Clermont quadruplex PCR phylotyping method and their antimicrobial susceptibility patterns.
This was a cross-sectional laboratory-based study carried out in the Clinical Microbiology and Molecular Biology Laboratories in the Department of Medical Microbiology, College of Health Sciences, Makerere University (Kampala, Uganda).
A total of 140 Uropathogenic E. coli strains that belonged to the bacterial collection of the Department of Medical Microbiology, College of health sciences Makerere University were studied. These strains had been isolated and stored at -80°C during a 12-month period (between January and December 2016). These isolates had been recovered from samples with a bacterial count of 105 CFU/ml of midstream urine samples of patients with a suspected UTI. The samples were selected by consecutive sampling and all samples that were poorly labeled and lacked traceable laboratory request form were excluded from the study. All isolates were plated on MacConkey agar and incubated at 37°C aerobically for 18–24 hours to obtain pure growth.
Antimicrobial susceptibility testing was done using the Kirby Bauer disk-diffusion method. Briefly, between one and five colonies of the isolate from the MacConkey agar were emulsified in saline and adjusted for turbidity to obtain a 0.5 McFarland standard. A sterile cotton swab on a stick was dipped in the colony-saline mixture, excess saline was then squeezed out by pressing the swab against the test tube, and the cotton swab gently applied onto the surface of the Mueller-Hinton-2- agar to obtain a uniform lawn. Up to six antibiotic impregnated discs were gently placed on the agar surface, at a minimum distant of 25 mm from each other, and the plates incubated at 37°C aerobically for 18–24 hours. The zones of inhibition diameters around each disc were measured using a ruler and compared against the zone diameter interpretative standards according to CLSI (2014). A panel of 12 antibiotics were used: ampicillin (30 μg), cefuroxime (30 μg), amoxicillin/clavulanic acid (20/10 μg), gentamycin (10 μg), trimethoprim/sulphamethoxazole (1.25/23.5 μg), chloramphenicol (5 μg), ciprofloxacin (5 μg), ceftriaxone (30 μg), ceftazidime (30 μg), imipenem (10 μg), nalidixic acid (30 μg) and nitrofurantoin (300 μg). E. coli ATCC 25922 was used as a control strain.
All isolates that showed an area of inhibition of diameter <22 mm for ceftazidime and <25 mm for ceftriaxone, which were selected for phenotypic detection of The test isolate was mixed in saline to obtain a suspension of 0.5 McFarland standard. The suspension was later swabbed to make a uniform lawn on Mueller-Hinton agar (MHA) plate. An amoxicillin-clavulanate (Augmentin) (20 μg/10 μg) disk was placed in the center of the plate with a 30-μg disk of a third-generation cephalosporin (ceftazidime and cefotaxime) at a distance of 20 mm from center to center on a Mueller Hinton agar (MHA) plate on opposite sides. The plate was later incubated aerobically at 37°C for 18–24 hours. All isolates that showed a clear extension of the edge of inhibition zone of the third-generation cephalosporin toward the augmentin disk were interpreted as positive for ESBL production.
Detection of carbapenemases was determined by a positive Modified Hodge test (MHT) as described by CLSI (2014); In brief, E. coli ATCC 25922 was streaked for confluent growth on Mueller-Hinton II agar plates. A disk saturated with 10 μg of imipenem was placed in the center of the plate, and each sample was then subsequently streaked from the disk to the edge of the plate. The presence of a distorted inhibition zone after overnight incubation was interpreted as a positive result. Klebsiella pneumoniae ATCC BAA-1705 and K. pneuomniae ATCC BAA- 1706 were used as positive and negative controls, respectively. Phenotypic detection of Metallo-beta-lactamases (MBL), K. pneuomniae producing carbapenems (KPC) and AmpC was carried out as described by Andrea Bartolini et al and Tsakris et al (Bartolini et al., 2014)(Tsakris et al., 2008).
DNA for amplification was extracted from whole cells by the boiling lysis method, as explained briefly below. A full loop of pure colonies from fresh pure cultures was suspended in 1 ml of sterile distilled water. The cells were lysed by heating at 95°C for 10 minutes. The cells were then vortexed for 5 seconds. Centrifugation was later done at 13,000 rpm for 5 minutes at room temperature and the sample kept at -20°C to harvest the supernatant containing the DNA. The supernatant was subsequently used for PCR as template DNA. The integrity of extracted DNA was evaluated by electrophoresis on a 1% agarose gel. The purity of DNA was also determined by the ratio A260/A280 using a spectrophotometer.
The distribution of phylogenetic groups amongst E. coli isolates was determined by the New Clermont Quadruplex PCR phylotyping method of 2013 (Clermont et al., 2013). Briefly, a single reaction mixture contained 2 μL of 10x buffer (supplied with Taq polymerase), 2 μL of DNA (approximately 100 ng), 20 pmol of each appropriate primer (except for AceK.f (40 pmol), ArpA1.r (40 pmol), trpBA.f (12 pmol), and trpBA.r (12 pmol)) (Shanghai Generay Biotech Co., Ltd.), 2mM of each dNTP, and 2U of Taq DNA polymerase (Fermentas, Lithuania) in a total volume of 20 μL. Primer sequences for the new Clermont’s quadruplex PCR phylogroup assignment method are as shown in Table 1. PCR amplifications were carried out on a thermal cycler Master-cycler gradient (Eppendorf, USA) under the following conditions: initial denaturation at 94°C for 4 min and 30 cycles for each denaturation at 94°C for 5 sec, annealing at 57°C for 20 sec (group E) or 59°C for 20 sec (quadruplex and group C), amplification at 72°C for 1 min, and final extension at 72°C for 5 min. PCR products were analyzed by electrophoresis with Qiaxel machine and a 2% agarose gel, stained with DNA safe stain (CinnaGen, Tehran, Iran) and visualized usingGelDoc 2000 transilluminator (Bio-Rad Laboratories, Milan, Italy). A molecular weight standard (100-bp ladder, Fermentas, Lithuania) was used.
The data was entered in an excel spreadsheet, cleaned and double-checked for missing variables duplicate entries and values out of range. The data was then exported to STATA version 14 for statistical analysis. Means and proportions were obtained. Chi-square test or the Fisher exact test was applied to compare categorical variables. P values < 0.05 were considered to be statistically significant.
Ethical approval (SBS-HDREC- 487) and a waiver of consent were sought to use stored clinical isolates from the higher degrees Research and Ethics Committee of the School of Biomedical Sciences, College of Health Sciences Makerere University and the Uganda National Council of Science and Technology.
Of the 140 E. coli isolates, 102 (72.9%) were females and 38 (27.1%) were males. The age of the patients was ranging from 2 to 91 years. The mean age was 36.27 with a standard deviation of 18.98.
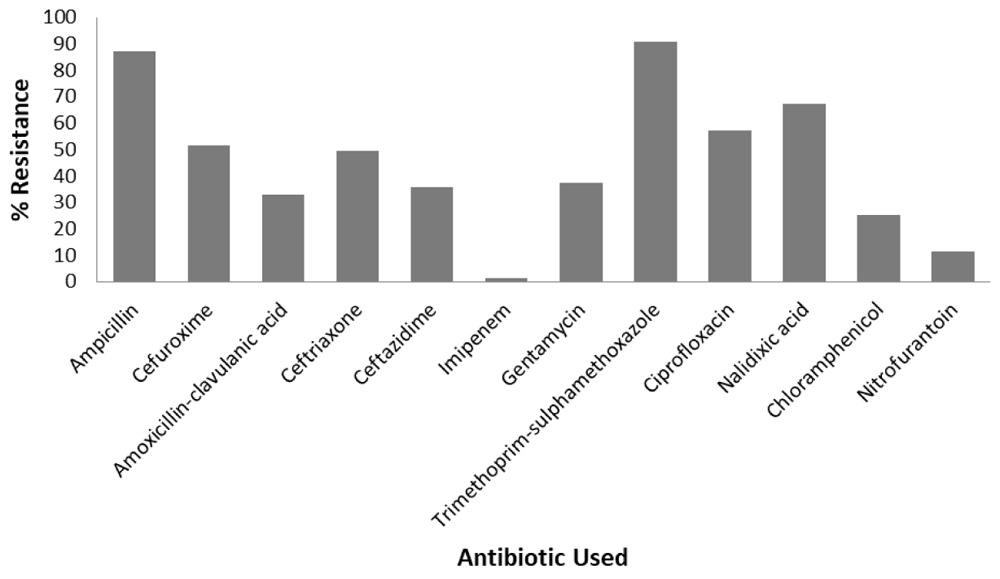
Resistance was highest to trimethoprim-sulphamethoxazole 127/140 (90.71%) followed by ampicillin 122/140 (87.14%) while resistance to nitrofurantoin 16/140 (11.43%) and imipenem 2/140 (1.43%) was minimal. Resistance to other antibiotics were as follows; cefuroxime 72/140 (51.43%), amoxicillin-clavulanic acid 46/140 (32.86%), gentamycin 52/140 (37.14%), chloramphenicol 35/140 (25%), ciprofloxacin 80/140 (57.14%), ceftriaxone 69/140 (49.29%), ceftazidime 50/140 (35.71%) and nalidixic acid 94/140 (67.14%). Figure 1 contains a bar chart depicting percentage resistance to different antibiotics. Data from disc-diffusion assays are available as Underlying data (Katongole, 2019).
In the study, 103/140 isolates (73.57%) were found to be MDR. In addition, 4/140 isolates (2.85%) were resistant to all antibiotics tested.
In this study, 61/140 (43.57%) were positive for ESBL production. Out of all the ESBL positive isolates, (57/61) 93.44% were MDR. All isolates were negative for carbapenem production. Only 2/140 (1.43%) were AmpC producers.
Phylogenetic group B2, in 56/140 of samples (40%), was the most prevalent, followed by group A in 9/140 samples (6.23%), Clade I/II in 7/140 samples (5%). The least prevalent were groups F (1/140 samples, 0.71%) and C (1/140 samples, 0.71%) (Table 2). Of the total isolates, 41.43% were unknown. Phylogenetic groupings for each patient are available as Underlying data (Katongole, 2019).
Phylogenetic group B2 was the most frequently resistant (38.57%). Phylogenetic groups B1 and F were the least resistant with a recorded highest resistance rate of 0.71% (Table 3).
The clinical management of UTIs is becoming a major burden due to the emergence of MDR uropathogens (Shabbir et al., 2018)(Flores-Mireles et al., 2015). Currently, third-generation cephalosporins are the most commonly used drugs in the management of complicated and uncomplicated UTIs (Stiller et al., 2017)(Bonkat et al., 2017). This study aimed to identify the phylogenetic groups of UPEC clinical isolates based on the Clermont quadruplex PCR method and to assess the relationship between these phylogroups and antibiotic susceptibility patterns in Uganda. In this study majority of the E. coli strains belonged to B2 (40%). This was similar to other studies worldwide (Iranpour et al., 2015)(Basu et al., 2013)(Moreno et al., 2008)(Takahashi et al., 2006). Other studies have, however, the most predominant phylogenetic group was group A (Ejrnæs et al., 2011)(Zhao et al., 2015). Phylogenetic group B2 has been associated with MDR-UPEC strains and increased expression of virulence factors (Ochoa et al., 2016)(Molina-López et al., 2011)(Nüesch-Inderbinen et al., 2017). Persistent and recurrent UTIs have also been associated with phylogenetic group B2 and this has been implicated in the pathogenesis of pyelonephritis(Ejrnæs et al., 2011)(Kudinha et al., 2013)(Luo et al., 2012). A similar study by Ramos et al. of pregnant mothers at Mulago National Referral Hospital found B1 to be the most predominant phylogenetic group (Ramos et al., 2012). The difference could be explained by the different study populations. In our study we used isolates from significant bacteriuria while in the study by Ramos et al., they used urine with asymptomatic bacteriuria(Ramos et al., 2012). Other studies have also demonstrated phylogenetic group B1 to be associated with commensal and less virulent strains of E. coli (Clermont et al., 2013)(Tenaillon et al., 2010). We also noticed the existence of phylogenetic groups A, C, E, F and clade I/II, which could not be detected using the triplex PCR method. This was similar with other previous studies(Iranpour et al., 2015)(Clermont et al., 2013)(Clermont et al., 2015)(Massot et al., 2016). The study also recorded 41.3% of the isolates that belonged to the unknown group; this could be due to new strains or the un-typable by PCR as explained by Clermont et al. (Clermont et al., 2013).
This study demonstrated very high rates of resistance to commonly used antibiotics, especially ampicillin, trimethoprim/sulphamethoxazole, and ciprofloxacin. This finding was similar to that of other studies(Gupta et al., 2001)(Kothari & Sagar, 2008). This could also be explained by the poor antimicrobial use policy in the country evidenced by the over the counter antimicrobial prescriptions(Mukonzo et al., 2013). We demonstrated increased prevalence of ESBL producing UPEC strains (43.57%). This was similar to other studies in the region(Kateregga et al., 2015)(Ampaire et al 2017). This could be explained by the increased pressure on third-generation cephalosporins in the management of UTIs(Ullah et al 2009)(Brosh-Nissimov et al., 2018).
In this study, we did not find any carbapenemases on phenotypic and genotypic screening. This was, however, different from other studies conducted in a similar setting(Okoche et al., 2015)(Kateete et al., 2016).
This study showed that group B2 isolates were the most resistant to most antimicrobials (38.57%). This was similar to other similar studies (Iranpour et al., 2015)(Massot et al., 2016). In addition, groups D, B1, and F were least resistant, again similar to other studies(Iranpour et al., 2015)(Massot et al., 2016).
In conclusion, our findings showed that group B2 (40%) were the most predominant and most resistant phylogenetic group among UPEC clinical isolates. About 9 % of E. coli isolates belonged to the newly described phylogroups C, E, F, and clade I. We recommend routine surveillance of antibiotic resistance patterns in the region to help clinicians make the treatment options for patients with UTIs. We recommend a longitudinal study employing whole-genome sequencing of E. coli strains in relation to UTI acquisition this will provide more insight on the role of Phylogenetic groups in the pathogenesis Of UTIs.
Figshare: Supplemental files_Paul Katongole et al_v2. https://doi.org/10.6084/m9.figshare.10006418.v1 (Katongole, 2019).
This project contains the disc-diffusion antibody sensitivity data and phylogenetic group for samples taken from each patient in this study.
Data are available under the terms of the Creative Commons Zero "No rights reserved" data waiver (CC0 1.0 Public domain dedication).
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Microbiology and molecular biology
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Partly
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Bacterial pathogenesis
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 30 Oct 19 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)