Keywords
Direct PCR, Mycoplasma genitalium
Direct PCR, Mycoplasma genitalium
Mycoplasma genitalium is an emerging sexually transmitted disease that was first identified and isolated in 19801 from men with non-gonococcal urethritis (NGU). Its epidemiology in connection to other STI syndromes been established since nucleic acid amplification assay development in the early 1990s2. The bacteria have been detected in substantial amounts from men with urethritis and women with cervicitis3. M. genitalium prevalence in the general population has been studied and found to be ranging between 1–3%4.
M. genitalium is found in roughly 15% of men with NGU and in 22% of men with non-chlamydial NGU. However, the associated infections do not have unique clinical symptoms, making it difficult to use clinical signs as a mode of identification5. Cervicitis has been described as the female version of male urethritis. M. genitalium is found in 10% of women with cervicitis6. Chlamydial coinfections in women with cervicitis are also common in some settings7. M. genitalium is a very fastidious bacterium and culturing of the bacterium is exhaustive and time consuming.
The introduction of polymerase chain reaction (PCR) assays has provided the necessary data for its clinical prevalence8. Many assays have been developed for the detection of M. genitalium in human specimens8–17. Most of these assays are mainly based on the PCR detection technique. Use of these PCR tests have shown that the disease spectrum is similar to those caused by Chlamydia trachomatis and Neisseria gonorrhoeae in both males and females13. However, these assays differ in their target DNA sequences, specimen preparation and amplicon detection methods. Many of these detection methods target the 16S rRNA and the MgPa protein genes. Conventional and, more recently, real-time PCR assays have been applied. Most of the detection studies have been conducted in the U.S.A, Europe and Australia, with various strains being discovered. In line with the detection of the bacterial species and its related infections in Africa, more studies need to be conducted on possible strains and their epidemiology. Whether the bacteria have links with other sexually transmitted infections can also be investigated. In this study, it is shown that direct PCR can be applied to the detection of M. genitalium from crude DNA extracts. M. genitalium prevalence and characteristics among female sex workers have been studied in Kenya and Uganda18–21. Its prevalence has also been studied among males who underwent circumcision in order to prevent HIV acquisition in Kisumu, Kenya22. However, most of these studies have focused on conventional, real-time PCR or transcription-mediated amplification assays for the detection of M. genitalium. This study reports the use of direct PCR for M. genitalium detection from crude DNA extracts using specific primers that target the 16S rRNA gene.
This study was approved by the Jomo Kenyatta University of Agriculture and Technology Institutional Ethics Review Committee (JKUAT-IERC): reference number JKU/2/4/896B. The swab samples were collected with written informed consent for the performance of further analysis.
The samples used in this study were collected as part of the sex workers outreach program (SWOP) central business district clinic in Nairobi, Kenya. As part of this program, patients who showed STI symptoms and consented to the study were sampled by taking vaginal swabs. The specimens were then put into sterile containers and transported to the Pan Africa Hub Laboratory (NUITM-KEMRI) within an hour and stored at -80°C. Anonymized samples were retrieved for use in this study.
The 352 swab lysates were prepared using the MightyPrep reagent for DNA (TAKARA BIO INC, Kusatsu, Shiga Prefecture, Japan; Cat No: 9182) using the manufacturer’s protocol with a slight modification. Swabs were cut and put into 1.5ml Eppendorf tubes. A total of 200uL of the MightyPrep reagent was added to the tubes and later centrifuged at 15krpm for one minute. The tubes were then transferred to a heated block at 95°C with shaking at 800rpm for 15 minutes. Later, the tubes were cooled down by lowering the heat block temperature to 25°C, followed by hard vortexing of each tube for one minute and, finally, centrifugation at 15krpm for two minutes before storage at -31°C.
The master mix was prepared using the manufacturer’s protocol with slight modifications (Hotstar Taq® Master Mix Kit 2.5 Units, Qiagen; Cat No: 203443). 100μM primer concentration was achieved by adding 303μl and 353μl of Tris EDTA (Nippon Gene Company Ltd, Japan, Cat No: 314-90021; 10mM Tris-HCl [pH 8.0], 1mM EDTA [pH 8.0]) to the forward and reverse primers (Table 1; Sigma-Aldrich, Darmstadt, Germany), respectively. The primers (Table 1) targeted the 16S rRNA gene23 giving 433bp amplicon size fragments. Master mix components were: RNase- free water (1x = 7.84μl); primer mix (1x = 0.08μl, 0.2μM of each primer); and HotStar Taq® master mix (2x), comprised of 2.5 units HotStarTaq DNA polymerase (1x = 10μl),1x PCR buffer (1x, contains 1.5mM MgCl2) and 200μM of each dNTP.
| Forward primer | Reverse primer |
|---|---|
| MG16-45F (TACATGCAAGTCGATCGGAAGTAGC) | MG16-447R (AAACTCCAGCCATTGCCTGCTAG-3’) |
After vortexing the master mix for five seconds, 18μl was aliquoted to each of the labeled 96 PCR tubes. 2μl of the swab lysates was added to each tube to make a final reaction volume of 20μl and the tubes were finger tapped for five seconds to mix the contents. A positive control (M. genitalium positive sample) and negative control (PCR water) were used. The PCR tubes were placed in the SimpliAmp™ Thermal Cycler (Applied Biosystems) and run under the following reaction conditions.
An initial antibody inactivation step was carried at 95°C for 15 minutes, followed by 35 cycles of: denaturation at 94°C for 60 seconds, annealing at 67°C for 60 seconds and extension at 72°C for 60 seconds. A final extension step was carried out at 72°C for 10 minutes, followed by the final hold at 4°C for ∞.
The PCR products were subjected to gel electrophoresis using 2.5% agarose gel SeaKem® GTG® agarose (Lonza, Rockland, ME, USA; Cat No: 50074) at 100V for 45 minutes. 6x loading dye (Nippon Gene; Cat No: 314-90261) was diluted with sample to make 1x and loaded onto the gel. The 100bp GelPilot® Ladder marker (Qiagen; Cat No: 239035) was used. The gels were stained with 2x GelRed™ Nucleic Acid Gel Stain (Biotium; Cat No: 41003) for one hour on a shaker. The image was viewed using the UltraSlim UV Transilluminator (Maestrogen).
The PCR products were subjected to another PCR. Master mix components were as described above. 18μl was aliquoted into the PCR tubes. 1μl of the sample products was added to the tubes to make a 19 μl final volume. The PCR tubes were placed in the SimpliAmp™ Thermal Cycler (Applied Biosystems) and run under the following reaction conditions.
An initial antibody inactivation step was carried out at 95°C for 15 minutes, followed by 30 cycles of: denaturation at 94°C for 60 seconds, annealing at 69°C for 60 seconds and extension at 72°C for 60 seconds. A final extension step was carried out at 72°C for 10 minutes, followed by the final hold at 4°C for ∞.
The products were run on a 2.0% agarose gel at 100V for 40 minutes. A 3000bp ladder (Solis BioDyne, Tartu, Estonia) was used. The gels were stained using 2x GelRed for one hour and viewed under an UltraSlim UV Transilluminator.
A total of 352 lysates were analyzed in this study. The results show evidence for the presence of M. genitalium from swabs taken from the female sex workers who were sampled. 352 lysates were prepared using the MightyPrep reagent.
M. genitalium was detected among the 352 swab lysates. Examples of M. genitalium detection are shown in Figure 3 to Figure 4. Figure 1 shows clear bands at positions 1, 2, 9, 10 and 17 on a 26-well agarose gel. The same kind of bands can be seen in Figure 2 at positions 9, 10, 11 and 18. The positive and negative controls are at positions 24 and 25, respectively, on each gel. A 600bp ladder was used at positions 1 and 26 to track the M. genitalium amplicon sizes of interest.
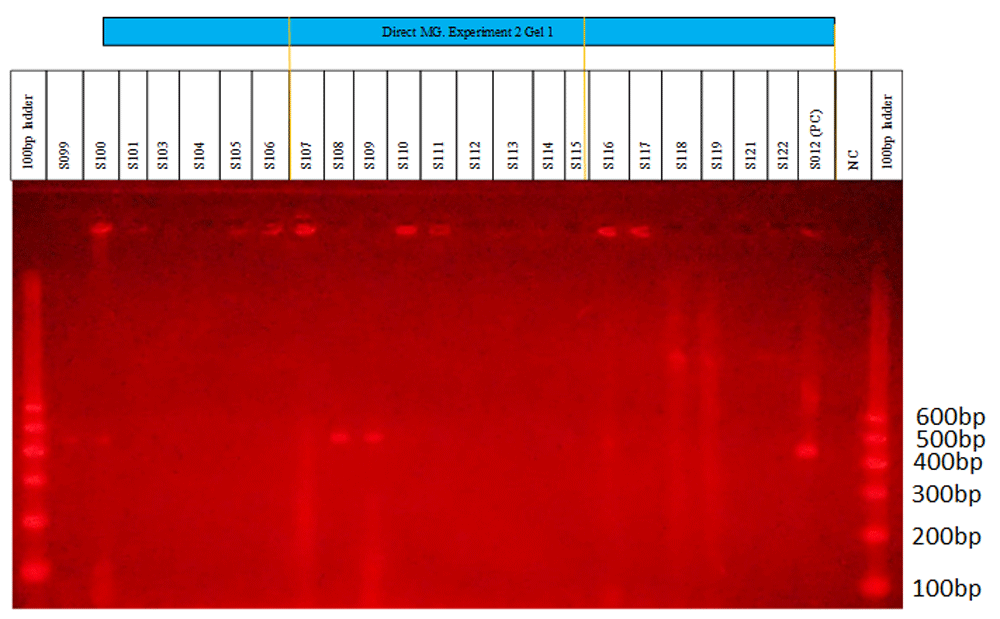
Ultraviolet camera gel image showing 22 samples run on a 26-well gel. The ladder is at positions 1 and 26 (Gel Pilot®). Clear Mycoplasma genitalium positive bands can be seen at positions 1, 2, 9, 10 and 17 (Sexually transmitted infection lysates S099, S100, S108, S109 and S116, respectively). The positive control (PC) and negative control (NC) are at positions 24 and 25, respectively.
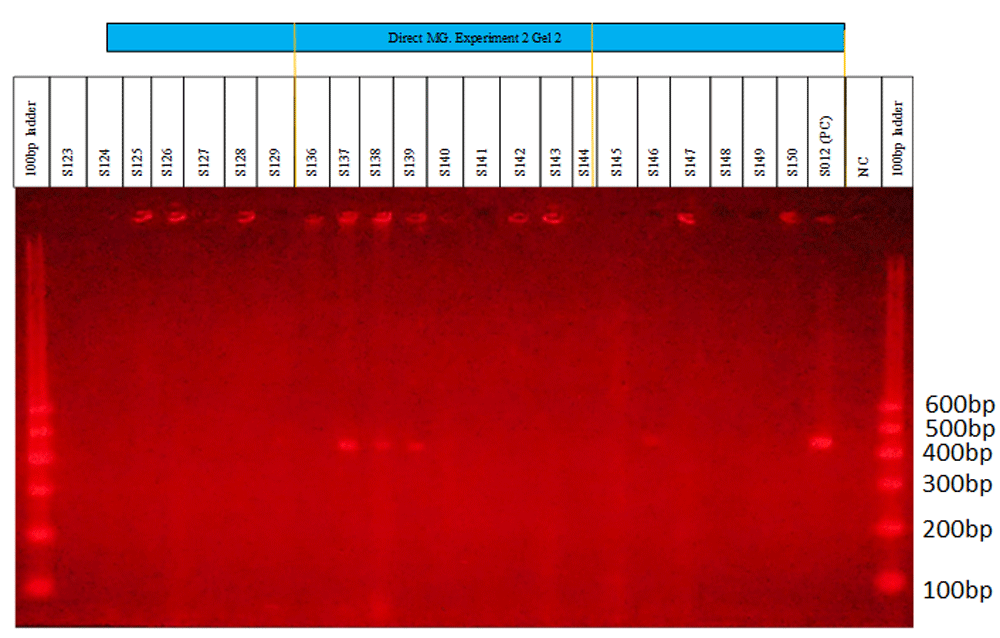
UV camera gel image showing the 22 samples run on a 26-well gel. The first and last wells represent the ladder (GelPilot®). Clear Mycoplasma genitalium positive bands can be seen at positions 9, 10, 11 and 18 (lysates S137, S138, S139 and S146, respectively). The positive control (PC) is shown at position 24 and the negative control (NC) at position 25.

The selected PCR products were amplified: the above image shows the first nine PCR products run on a gel after the amplification was conducted. The ladder (Solis BioDyne) is at the first and last lanes. The positive control (PC) is at lane 11, while the negative control (NC) is at lane 12.

This gel shows PCR products of the samples that were selected for amplification. The first and last lane contain the ladder marker (Solis BioDyne), the PCR products run from lanes 2 to 10 and the positive control (PC) is at position 11, while the negative control (NC) is at position 12.
The PCR products were subjected to another amplification reaction. After the reaction, the products were run on a 13-well agarose gel. A 3000bp ladder was loaded on positions 1 and 13, with the positive and negative controls at positions 11 and 12, respectively, as can be seen in Figure 3 and Figure 4. Out of the 352 swab lysates used, 29 tested positive for M. genitalium.
Recently, DNA amplification protocols using PCR have been employed in the detection of M. genitalium. To investigate the presence of M. genitalium from the clinical swab samples collected from a clinic for sex workers in Nairobi, Kenya, a direct PCR technique was used for the detection of M. genitalium. This technique involves the use of target-specific primers to select the DNA of interest from a crude extract. The study detected M. genitalium using primers that bind to the 16SrRNA gene from the crude DNA extract. Jensen and his colleagues23 developed a wide range of primers that target the 16S rRNA gene, producing different amplicon sizes. A novel PCR was used24 to detect M. genitalium using oligonucleotide primers that corresponded to sequences along its 16S rRNA gene.
The study was able to detect 29 M. genitalium positive samples out of the 352 lysates. However, the challenge experienced with this method was non-specific amplification, realized from the multiple fragments produced. A possible solution to this is in the use of more precise target-specific primers to prevent the amplification of genes with closely related sequences. Application of this method can be a remedy to the constant loss of DNA due to long extraction processes, at the same time maintaining its quality for further downstream analysis.
M. genitalium prevalence was shown to be at 8.24%. This shows that one out of every eight patients sampled was positive for M. genitalium related infections. Balkus and colleagues in 2018 were able to detect M. genitalium from 25 out of 221 (11.3%) women from Kenya and the US25. Prevalence rates of 12.9%18 and 16%19 have also been reported among sex workers in Nairobi, Kenya. The prevalence obtained in this study therefore does not show any significant drop in M. genitalium infections. Despite better and improved access to healthcare, M. genitalium infections seem to continue to be a burden. Possible reasons might be due to having multiple sex partners26 or antibiotic resistance to drugs of choice such as macrolides and fluoroquinolones27,28
Overall, the prevalence results suggest that more measures need to be taken to control M. genitalium infections. Awareness campaigns need to be carried out to sensitize people on preventive measures rather than taking potential risks that may lead to exposure to the infection. Studies need to be done to investigate M. genitalium drug resistance. This will be helpful in informing policy and practice. As a result, screening can be done in patients to check for resistance before prescribing medication.
Figshare: Detection of Mycoplasma genitalium using Direct PCR. https://doi.org/10.6084/m9.figshare.10282691.v129.
This project contains the following underlying data:
- Direct PCR 1.docx – Direct PCR 4.docx (lists of the samples tested for Mycoplasma genitalium in four sets of 88 samples)
- Exp 1 Gel 1.JPG - Exp 4 Gel 4.JPG (gel electrophoresis of PCR products; 100bp GelPilot® Ladder marker [Qiagen] at positions 1 and 26, samples from positions 2 to 23, positive control at position 24 and negative control at position 25)
- Amplification Gel 1.JPG - Amplification Gel 3.JPG (gel electrophoresis of the amplified products; 100bp ladder [Solis BioDyne] at positions 1 and 13, samples from positions 2 to 10, positive control at position 11 and negative control at position 12)
- Amplification Gel 4.JPG (gel electrophoresis of the amplified products; 100bp ladder [Solis BioDyne] at positions 1 and 13, samples from positions 2 to 7, positions 8, 11 and 12 contain no samples [blanks], positive control at position 9 and negative control at position 10).
Data are available under the terms of the Creative Commons Zero "No rights reserved" data waiver (CC0 1.0 Public domain dedication).
The authors would like to thank the Nagasaki University Institute of Tropical Medicine in collaboration with the Kenya Medical Research Institute (NUITM-KEMRI), The Pan Africa University Institute of Science Technology and Innovation hosted at the Jomo Kenyatta University of Agriculture and Technology, Kenya (PAUSTI-JKUAT) for the research conceptualization and support.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Not applicable
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: molecular biology, microbiology
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Not applicable
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Infectious diseases and microbiology, epidemiology especially in the area of Neisseria sp and STI.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 26 Nov 19 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)