Keywords
Text-mining, Cancer
Text-mining, Cancer
Cancer is a genetic disease1, with relevant genes often identified through functional screening2–4, gene expression profiling5–7, or genomic sequencing experiments8–10. While researchers may need to quickly understand the published literature regarding a particular gene, the large volume of publications can make this challenging. Indeed, a search of PubMed finds >582,000 citations with the keywords “cancer” and “gene”, with >175,000 articles published within the previous 5 years. The large volume of cancer genomic publications necessitates the development of tools to help cancer researchers navigate and summarize articles efficiently.
The use of controlled vocabularies and text-mining tools has facilitated the annotation and searching of biomedical literature. In particular, the National Library of Medicine’s Medical Subject Headings provides a controlled vocabulary of MeSH terms for indexing MEDLINE/PubMed articles. PubTator is a web-based platform, designed to assist biocuration, that uses robust text-mining tools to annotate PubMed articles with respect to genes, chemicals, species, and mutations11. However, while PubTator allows users to search PubMed based on these biological concepts, summaries of the results are not provided. Other tools such as Anne O’Tate12 and PubReminer13 summarize PubMed searches, but are not cancer-specific and have limitations regarding the number of results that can be returned. Anne O’Tate, for example, allows users to search PubMed and summarizes the results based on important words, phrases, authors, and other fields12. PubReminer13 also allows PubMed queries and summarizes articles based on common words, MeSH terms, and other fields. These summaries are useful but are not cancer specific, and cancer type mentions can be difficult to find and may not appear in the search results.
Here we develop a Cancer Publication Portal (CPP), a web application to help users search and summarize the cancer genomic literature14,15. CPP allows a user to enter a human gene or gene set of interest, and then summarizes the relevant cancer-related publications mentioning this gene through tabular and graphical summaries showing the frequency of articles by cancer type, pharmacological agent, genomic mutation, and additional human genes mentioned in article titles or abstracts. Additionally, CPP catalogs and summarizes articles based on mentions of >30 cancer-related terms. The tool is designed to provide users with publication statistics, such as the number of articles mentioning EGFR and erlotinib across cancer types, as well as to facilitate exploration and retrieval of relevant articles. For example, a user can quickly find all articles based on a set of genes and a collection of cancer types. Interactive summaries allow users to narrow in on a topic of interest by applying additional filters. Results are summarized across cancer types, and the process can be repeated. At any point, the user can access article abstracts from PubTator, as well as download statistical summaries. In this fashion, researchers and students can explore the literature and find articles in a gene- and cancer-focused way.
CPP14,15 integrates data from a variety of sources including PubTator11, the Medical Subject Headings (MeSH) database; the HUGO Gene Nomenclature Committee (HGNC) human gene name database16, the PubMed database of biomedical literature citations, and the National Cancer Institute (NCI) Thesaurus17. An overview of data collection is provided in Figure 1A. Generally, article association data for genes, chemicals, diseases, and mutations was collected from PubTator, then updated and filtered as described below and in Figure 1A. Cancer term associations were identified by comparing PubMed abstracts, as described below and in Figure 1A. Summary statistics for CPP, which contains article-entity association information for 1,143,191 articles and 19,551 genes, is provided in Table 1. The mean number of articles per gene is 138.6 (median = 9.0), but the number of publications per gene is uneven, with the 192 most frequently mentioned genes accounting for >50% of all publications. The three most frequently mentioned genes are TNF (N = 64,103), TP53 (62,602), and EGFR (35,178) (Extended data, Supplementary Table S1)18. The three most common cancer types are Breast Neoplasms (N = 119,891), Leukemia (N = 103,222), and Lymphoma (N = 60,320) (Extended data, Supplementary Table S2)18.

(A) CPP integrates data from PubTator, PubMed, HGNC, MeSH, and the NCI Thesaurus to summarize articles based on their references to cancer type, mutations, genes, and cancer-related terms. (B) Selected cancer-related terms identified from the titles/abstracts of ~3.5 million publications. The log10 ratio of the cancer publication frequency to non-cancer publication frequency is shown.
| Variable | Frequency | |
|---|---|---|
| Entity | Article-entity associations | |
| Genes Cancer types Cancer terms Pharmacological agents Mutations Articles | 19,551 660 37 5,565 32,854 1,143,191 | 2,710,512 1,626,382 4,571,767 726,501 91,831 - |
Collection and processing of PubTator data. Data defining article-gene, article-disease, article-chemical, and article-mutation relationships were downloaded from PubTator via FTP. PubTator data defines associations between articles and mentions of genes, chemicals, diseases, and mutations in article titles or abstracts. MeSH terms for descriptor and supplemental record sets were downloaded as XML files and parsed to extract MeSH IDs and their corresponding terms. A list of pharmacologically active compounds was also downloaded from MeSH via its FTP service. PubTator data was filtered to include only those articles mentioning human genes and only those articles that are cancer-related, i.e., that mention MeSH terms falling under the heading “Neoplasm” (Tree Number C04). We take advantage of MeSH tree structure and remove redundant MeSH IDs if a child MeSH ID is mentioned in the same article. For example, an article mentioning both “Breast Neoplams” (C04.588.180) and “Triple Negative Breast Neoplasms” (C04.588.180.788) would have the former removed. We also recode “Neoplasms” (C04) as “Neoplasms (unspecified)” (C04.000) if this is the only cancer MeSH term associated with the article. Obsolete MeSH IDs were updated by testing the terms associated with that entry against current MeSH headings and Supplementary Concept record terms. Mutation data was reformatted according to HGVS sequence variant nomenclature recommendations19. For each mutation, we identify the gene or genes most commonly associated with it, and store this information in CPP.
Identification and annotation of cancer terms. In addition to the article associations provided by PubTator, we report associations between articles and “cancer terms”. We identified cancer-terms by comparing title/abstract word ‘stems’ between cancer-related (N=851,868) and non-cancer related articles (N=2,607,020) that mentioned at least one human gene. Abstracts were downloaded from PubMed’s FTP service. These titles/abstracts contained a total of 5,564 unique word stems, with 2,633 word stems more common in cancer articles (P < 0.01, Fisher’s exact test). In order to focus on word stems that would be most informative in a cancer-specific context, we filtered these results by considering only word stems that occurred in > 1% of cancer-related articles. Word stems related to disease/tissue (e.g., ‘renal’), gene name (e.g., ‘kinase’), and miscellaneous words (e.g., ‘report’) were also filtered out. Word stems for similar words were combined, based on common word usage and the NCI Thesaurus17. In addition, several terms deemed important by the authors (e.g., “immunotherapy”) were added, even if occurring < 1% of the time in cancer-related articles. The full list of 37 cancer terms can be seen in the Extended data, Supplementary Table S318. Selected terms are shown in Figure 1B.
For each cancer term, we find a non-redundant set of word stems corresponding to the term itself and its synonyms according to the NCI thesaurus (Extended data, Supplementary Table S3)18. Such an approach allows us to identify a concept (e.g., ‘mutation’) even when a synonym (e.g., ‘genetic alteration’) is used. For each cancer-related article, we search its title/abstract for mentions of cancer terms and add these relationships to the CPP database.
Technical details. Python and R v3.5.2 were used for data processing. PubMed files were parsed using the Python “PubMed Parser”20 and word stems found using the Snowball stemmer from Python’s NLTK module, after removal of stop words and any word with no more than 3 characters. Additional XML files were parsed using the Python module lxml. After processing, data was loaded into a MySQL database. The web interface was developed using R/Shiny v1.2.0.
CPP runs in a standard web browser and is available for public use at the following address: https://gdancik.github.io/bioinformatics/CPP/.
CPP14,15 takes a gene-centric approach for finding and summarizing cancer-related articles based on mentions of cancer types, cancer terms, drugs, mutations, and additional genes. In order to demonstrate the utility of CPP, we use CPP to summarize and explore cancer-related articles mentioning the gene epidermal growth factor receptor (EGFR), a gene mutated in >30% of patients with non-small cell lung cancer21 and a gene that can be targeted by tyrosine kinase inhibitors such as gefitinib and erlotinib22. The user starts by selecting the desired gene from the drop-down menu. After selecting EGFR, CPP tells us that there are 35,178 articles found, covering 422 cancer types (Figure 2A). We note that “cancer types” here is defined according to MeSH subject headings, which categorize cancers by both site and histological type. The top three cancer types are “Neoplasms, Glandular and Epithelial”, “Thoracic Neoplasms”, and “Lung Neoplasms”. We next select the cancer types to search, by either clicking on the table or selecting cancer types from the drop-down menu. Cancer types can also be uploaded from a file. After clicking on the button to retrieve the summaries, we get a tabular and graphical summary showing the number of articles mentioning both the selected gene and each selected cancer type (Figure 2B). If no cancer type is selected, then all cancer types will be summarized. Here it is easy to compare the number of articles mentioning EGFR across cancer types, and a user quickly sees that lung cancer is the most common.
Figure 3 shows additional summaries provided by CPP. Summaries include frequency tables showing the number of articles associated with the selected gene and Cancer Terms, Drugs, Mutations, and Additional Genes; and stacked bar graphs for visualizing the distribution of each entity across cancer types. If the user searched for multiple genes, a summary of the selected genes is also provided. The frequency tables allow a user to quickly identify entities (such as drugs) that are commonly mentioned in the literature, while the stacked bar graphs allow a user to qualitatively evaluate when entities are more (or less) associated with specific cancer types than others. In this example, frequency tables show that gefitinib and erlotinib are the two drugs most commonly associated with EGFR (Figure 3A), the EGFR mutations p.T790M and p.L858R are most common (Figure 3B), and ERBB2, KRAS, and TP53 are the genes that most frequently co-occur with EGFR (Figure 3C). However, in the latter case the stacked bar graph shows that these co-occurring genes are associated with specific cancer types. Specifically, while KRAS is the most common gene that co-occurs with EGFR in lung cancer, the most common genes that co-occur with breast cancer and glioma are ERBB2 and TP53, respectively (Figure 3C). Such associations may reflect genomic differences between cancer types, or may reflect a bias in the literature. The stacked bar graphs are interactive. A user can single click on an entity in the legend to hide the entity from the graph, and double-click on an entity to hide all other entities. This toggling can be canceled by clicking or double clicking the entity a second time. For example, by double clicking on the drug irinotecan, we can see that this drug is associated with colorectal cancers more than other cancer types (Figure 3A, inset).
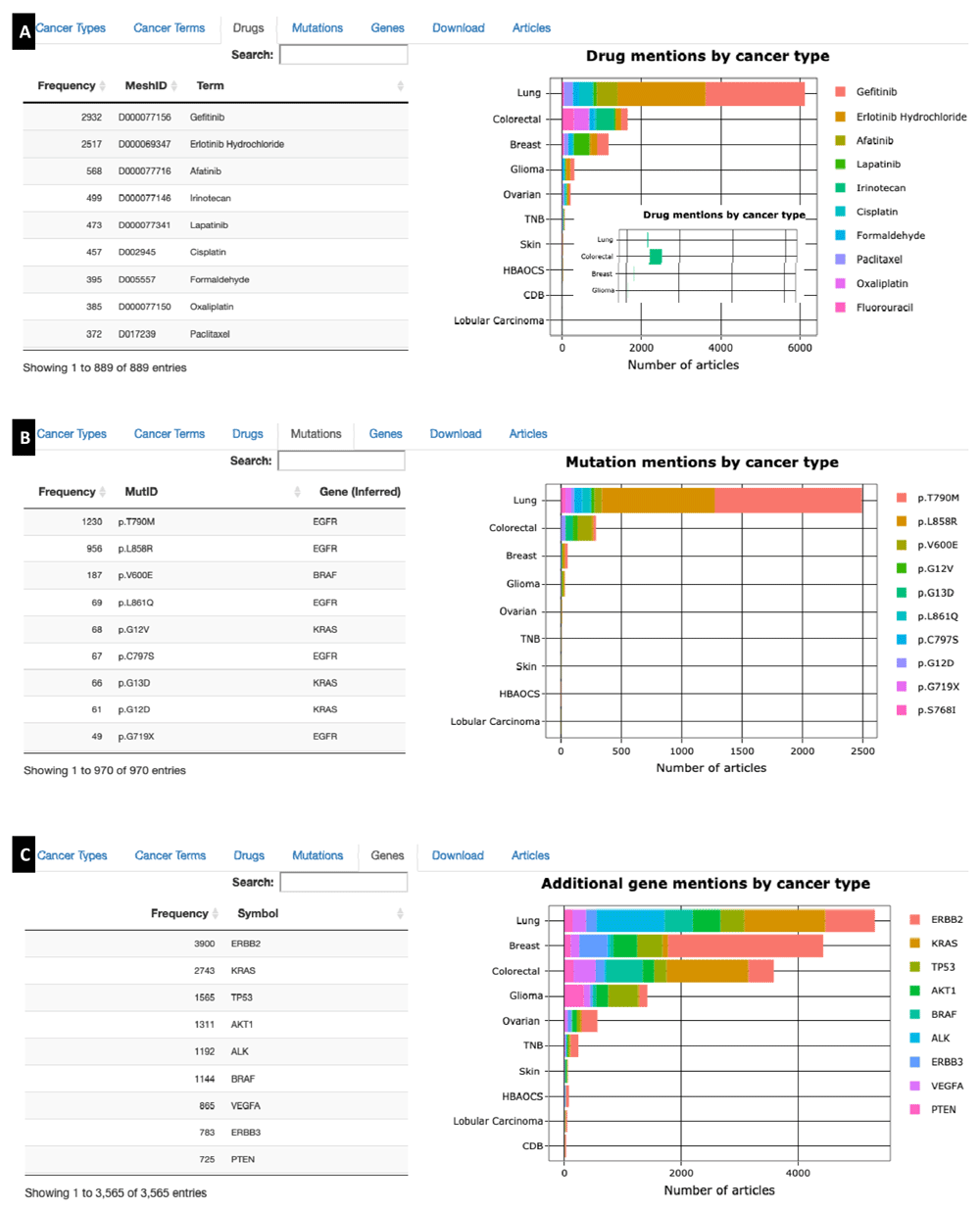
Summaries and stacked bar graphs are provided for Cancer terms (not shown), (A) Drugs, (B) Mutations, and (C) mentions of additional Genes. Inset in (A) shows only irinotecan, obtained by double clicking on that drug in the legend.
In addition to summarizing the cancer genomic literature, CPP is designed to help users explore the literature and quickly find articles of interest. After selecting one or more genes and cancer types, a user can add filters by clicking on one or more rows of any frequency table to add an entity to the filter. For each entity type, the user can retrieve articles that mention either all of the selected terms or any of the selected terms. For example, a user interested in publications assessing the predictive value of EGFR mutations as biomarkers for gefitinib or erlotinib treatment could use CPP to first find articles that mention EGFR and any cancer type. Then the user can specify additional filters to limit the results to articles that mention both mutation and survival, and either of the drugs gefitinib or erlotinib (Figure 4A). Note that cancer term filters are based on word ‘stems’ and synonyms from the NCI Thesaurus, and therefore will recognize variations of the search term. For example, the cancer term “mutation” includes any words with a stem of ‘mutat’, which includes the words “mutation”, “mutations”, and “mutated”; and word stems corresponding to ‘genetic alteration’ and ‘genetic change’ (Extended data, Supplementary Table S3)18. This search results in 1,169 articles being found. In summarizing the articles, we see that lung cancers are the dominant cancer type (Figure 4B), and that the number of articles is similar for each drug across cancer types (Figure 4C). We note that the stacked bar graphs are now limited to the entities we have selected (i.e., gefitinib and erlotinib).
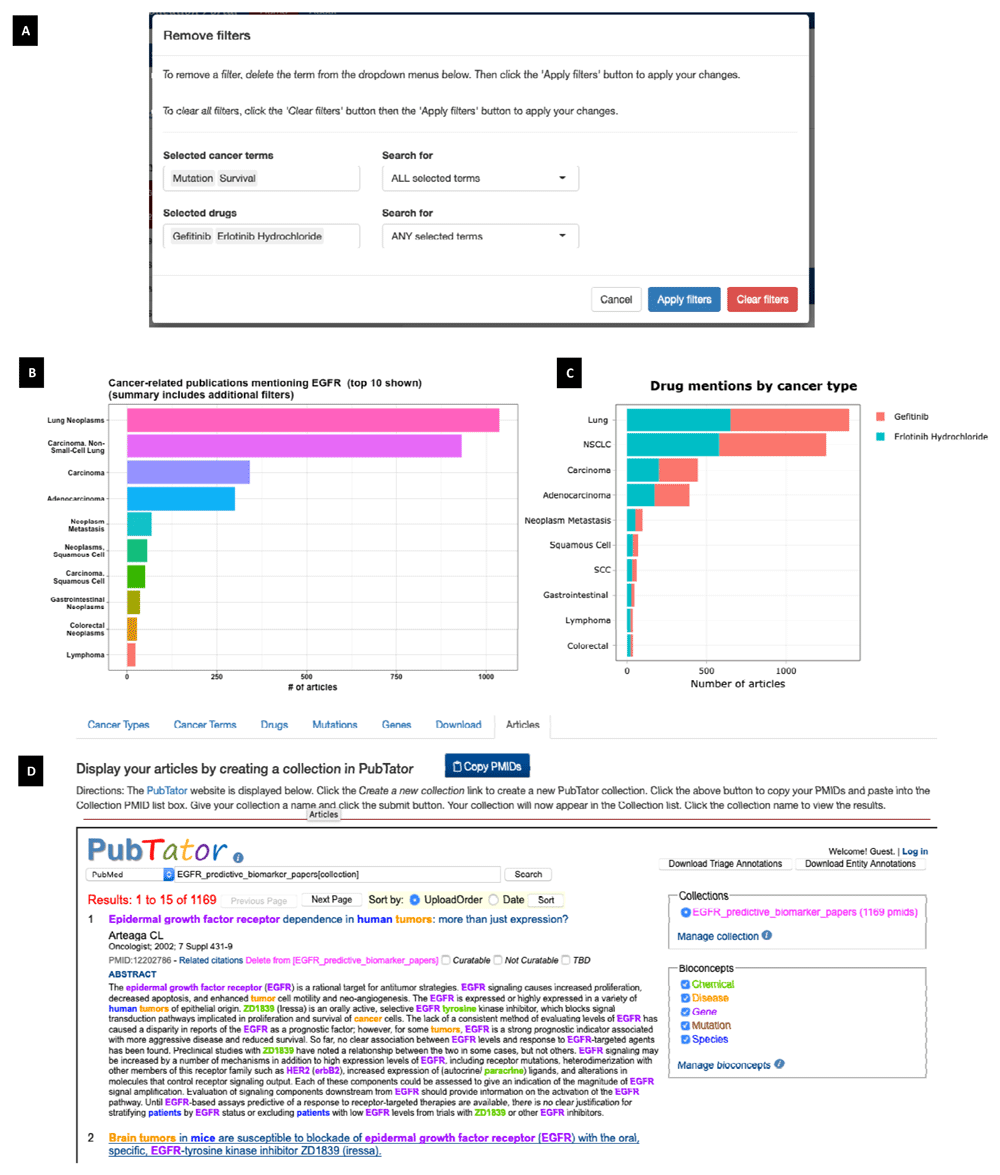
(A) Filters can be specified for all selected terms for an entity or any selected term. Current filter shows articles mentioning mutation and survival and either gefitinib or erlotinib. (B) Cancer summary of results for EGFR, all cancer types, and the filters in (A). (C) Stacked bar graph showing mentions of gefitinib and erlotinib across cancer types. (D) Screenshot of ‘Articles’ tab where user can create a PubTator collection to view the current set of articles.
Users can easily explore the literature by adding and removing filters. At any point, a user can view abstracts for the current set of articles. A user views abstracts by selecting the ‘Articles’ tab, clicking the ‘Copy PMIDs’ button, and then creating a new collection in PubTator, which is displayed on the Articles page using an iframe (Figure 4D). This allows a user to seamlessly view relevant abstracts after applying the desired filters. Additionally, a user can download results to a CSV file, for the current list of PMIDs, as well as frequency summaries for Cancer Types, Cancer Terms, Drugs, Mutations, and Additional Genes from the ‘Download’ tab.
CPP14,15 is designed to help users efficiently explore and summarize the cancer genomic literature, and should be useful for cancer researchers who are looking for relevant articles for a gene of interest, for meta-researchers who study the publication landscape, and for students learning about the relationship between one or more genes and cancer types. Because CPP quickly summarizes articles across cancer types, CPP can be used to assess whether a gene might be novel for a particular cancer type (or any cancer type), based on the frequency of gene mentions in titles/abstracts of cancer-related publications. CPP also provides summaries of cancer terms, drugs, mutations, and co-occurring genes that can connect a researcher to key biological concepts and the underlying articles that might inform their research. The use of cancer terms, unique to CPP, summarizes articles in a cancer-specific way and allows for quick retrieval of articles based on a cancer term, such as tumor suppressor, chemotherapy, or metastasis. Furthermore, because CPP identifies associations between articles and cancer terms by using the NCI Thesaurus and ‘stem’ words to cover synonyms and variant word forms (such as ‘mutation’ and ‘mutated’), CPP will retrieve a more valid set of articles than a simple PubMed search for a term in other databases.
Despite its utility, CPP has several limitations that are common in all text-mining based tools. With the exception of cancer terms, article associations in CPP are derived from PubTator associations. While some associations may be missed, PubTator uses cutting edge text-mining tools and the F1 scores for gene, disease, and mutation identification are all > 80%11.
Importantly, text-mining associations are determined only by textual relationships and may not reflect underlying biology. For example, genes that are mentioned together in the same abstract may or may not interact. Similarly, publication frequency may reflect publication bias and not biological importance. Other tools are available for looking at specific biological relationships, such as STRING for protein interactions23, and cBioPortal for Cancer Genomics for exploring genomic datasets24. CPP is designed to complement these tools by providing an overview of the cancer genomic literature and by helping researchers quickly find relevant publications. Finally, all CPP associations, which are derived from PubTator, are based on entity mentions in titles and abstracts only. However, the recently released PubTator Central includes associations from the PubMed Central (PMC) Text Mining Subset25. PMC contains the full text of ~3 million articles, though we expect <4% to be cancer-related. PubTator Central associations will be integrated into CPP in a future release.
In conclusion, CPP is an easy-to-use web application that allows researchers to efficiently summarize and search the cancer literature for articles based on one or more genes of interest. CPP will be updated approximately once a month following PubTator data releases.
Associations between genes, diseases, chemicals, mutations, and articles were downloaded from the PubTator FTP page (ftp://ftp.ncbi.nlm.nih.gov/pub/lu/PubTator/)
MeSH descriptor files were downloaded from https://www.nlm.nih.gov/databases/download/mesh.html
Dataverse: Cancer Publication Portal: an online tool for summarizing and searching human cancer-genomic publications (supporting data). https://doi.org/10.7910/DVN/BYKF1L18
This project contains the following extended data:
Supplementary Table S1. Number of articles per gene in Cancer Publication Portal.
Supplementary Table S2. Number of articles per cancer type in Cancer Publication Portal. Cancer types are defined by cancer-related MeSH TreeIDs (C04*).
Supplementary Table S3. Cancer terms and synonyms included in Cancer Publication Portal. For each term, there was a statistically significant difference in the proportion of mentions between cancer-related and non-cancer related articles (P < 0.001 by Fisher’s exact test). *, term is included despite appearing in < 1% of cancer-related articles, and/or not being cancer-specific (i.e., log10 ratio < 1).
Extended data are available under the terms of the Creative Commons Zero "No rights reserved" data waiver (CC0 1.0 Public domain dedication).
CPP is available as a web resource at: https://gdancik.github.io/bioinformatics/CPP/.
Source code for the web interface is available from: https://github.com/gdancik/CPP.
Source code for data processing and database creation is available from: https://github.com/gdancik/CPP_setup.
The database is available from the following docker image: https://hub.docker.com/r/gdancik/dcast.
Archived source code for web interface at time of publication: https://doi.org/10.5281/zenodo.355011014.
Archived source for data processing and database creation at time of publication: https://doi.org/10.5281/zenodo.355011215.
License: GNU General Public License-2.
The authors acknowledge Stefanos Stravoravdis for coding contributions, and Andrew Johnson for coding contributions and technical assistance. The authors also acknowledge Jason Duex and Sunny Guin for testing and providing feedback for an earlier version of the tool.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the rationale for developing the new software tool clearly explained?
No
Is the description of the software tool technically sound?
Yes
Are sufficient details of the code, methods and analysis (if applicable) provided to allow replication of the software development and its use by others?
Yes
Is sufficient information provided to allow interpretation of the expected output datasets and any results generated using the tool?
Yes
Are the conclusions about the tool and its performance adequately supported by the findings presented in the article?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Protein-protein interaction prediction, protein orthology, text-mining, statistical analysis of experimental data, web/tool development.
Is the rationale for developing the new software tool clearly explained?
Yes
Is the description of the software tool technically sound?
Partly
Are sufficient details of the code, methods and analysis (if applicable) provided to allow replication of the software development and its use by others?
Yes
Is sufficient information provided to allow interpretation of the expected output datasets and any results generated using the tool?
Yes
Are the conclusions about the tool and its performance adequately supported by the findings presented in the article?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Human genomics, genetics, comparative genomics, bioinformatics, nomenclature, biomedical resources.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 10 Dec 19 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)