Keywords
Evolution, Natural selection, Adaptation, Viruses, Codon Usage Bias, R, RStudio
Evolution, Natural selection, Adaptation, Viruses, Codon Usage Bias, R, RStudio
During the translation process from mRNAs to proteins, information is transmitted in the form of triple nucleotides, named codons, which encode amino acids. Multiple codons that encode one amino acid are known as synonymous codons. Studies concerning different organisms report that synonymous codons are not used uniformly within and between genes of one genome, a phenomenon known as codon usage bias (CUB)1,2. Since viruses rely on the tRNA pool of their hosts in the translation process, previous studies suggest that translation selection or/and directional mutational pressure act on the codon usage of the viral genome to optimize or deoptimize it towards the codon usage of their hosts3,4.
Tools and packages are available to analyze codon usage, e.g. coRdon5, but there is no package available that focuses on the examination of codon usage co-adaptation between viruses and their hosts. vhcub is a package implemented in R, which aims to easily analyze the co-adaptation of codon usage between a virus and its host. vhcub measures several codon usage bias measurements, such as effective number of codons (ENc)6, codon adaptation index (CAI)7, relative codon deoptimization index (RCDI)8, similarity index (SiD)9, synonymous codon usage orderliness (SCUO)10, and relative synonymous codon usage (RSCU)10. It also provides a statistical dinucleotide over- and under-representation with three different models.
vhcub imports Biostrings11, seqinr12 and stringr13 to handle fasta files and manipulate DNA sequences. Also, it imports coRdon5, which is used to estimate different CUB measures.
vhcub first converts the fasta format to data.frame type, to efficiently maintain and calculate different indices implemented in the package. Table 1 describes all the functions available in vhcub, and the result returned from each. Also, it contains references to the equations used to estimate each measure. Furthermore, vhcub uses ggplot214 to visualize two important plots named ENc-GC3 plot (Figure 2) and PR2-plot (Figure 3), which help to explain the factors influencing a virus’s evolution concerning its CUB.
| Function name | Description | Value |
|---|---|---|
| fasta.read | Read fasta formate and convert it to data frame | A list with two data.frames; the first one for virus DNA sequences and the second one for the host. |
| CAI.values | Measure the Codon Adaptation Index (CAI) using Sharp and Li (1987)7 equation, of DNA sequence. | A data.frame containing the computed CAI values for each DNA sequences within df.fasta. |
| dinuc.base | A measure of statistical dinucleotide over- and under-representation; by allows for random sequence generation by shuffling (with/without replacement) of all bases in the sequence13. | A data.frame containing the computed statistic for each dinucleotide in all DNA sequences within df.virus. |
| dinuc. codon | A measure of statistical dinucleotide over- and underrepresentation; by allows for random sequence generation by shuffling (with/without replacement) of codons13. | A data.frame containing the computed statistic for each dinucleotide in all DNA sequences within df.virus. |
| dinuc. syncodon | A measure of statistical dinucleotide over- and underrepresentation; by allows for random sequence generation by shuffling (with/without replacement) of synonymous codons13. | A data.frame containing the computed statistic for each dinucleotide in all DNA sequences within df.virus. |
| ENc.values | Measure the Effective Number of Codons (ENc) of DNA sequence. Using its modified version (Novembre, 2002)6. | A data.frame containing the computed ENc values for each DNA sequences within df.fasta. |
| GC.content | Calculates overall GC content as well as GC at first, second, and third codon positions. | A data.frame with overall GC content as well as GC at first, second, and third codon positions of all DNA sequence from df.virus. |
| RCDI.values | Measure the Relative Codon Deoptimization Index (RCDI)8 of DNA sequence. | A data.frame containing the computed ENc values for each DNA sequences within df.fasta. |
| RSCU. values | Measure the Relative Synonymous Codon Usage (RSCU)7 of DNA sequence. | A data.frame containing the computed RSCU values for each codon for each DNA sequences within df.fasta. |
| SCUO. values | Measure the Synonymous Codon Usage Eorderliness (SCUO) of DNA sequence using Wan et al., 200410 equation. | A data.frame containing the computed SCUO values for each DNA sequences within df.fasta. |
| SiD.value | Measure the Similarity Index (SiD) between a virus and its host codon usage15. | A numeric represent a SiD value. |
| PR2.plot | Make a Parity rule 2 (PR2) plot16, where the AT-bias [A3/(A3 +T3)] at the third codon position of the four- codon amino acids of entire genes are the ordinate and the GC-bias [G3/(G3 +C3)] is the abscissa. The centre of the plot, where both coordinates are 0.5, is where A = U and G = C (PR2), with no bias between the influence of the mutation and selection rates. | A ggplot object. |
| ENc. GC3plot | Make an ENc-GC3 scatterplot17. Where the y-axis represents the ENc values and the x-axis represents the GC3 content. The red fitting line shows the expected ENc values when codon usage bias affected solely by GC3. | A ggplot object. |
vhcub was developed using R and is available on CRAN. It is compatible with Windows, and major Linux operating systems. The package can be installed as:
install.packages( "vhcub" )
Figure 1 describes the vhcub workflow. It starts with reading the fasta files for a virus and its host. After, nucleotide content analysis, codon usage bias analysis on genes and codon level (marked by the red boxes in Figure 1) can be applied independently (the blue boxes in Figure 1). However, within the same analysis, some measures rely on others. For example, the reference set of genes used to estimate a virus codon adaptation index was defined based on the effective number of codons of its host. Finally, the orange boxes in Figure 1 represent the two plots (ENc-GC3 plot and PR2-plot).
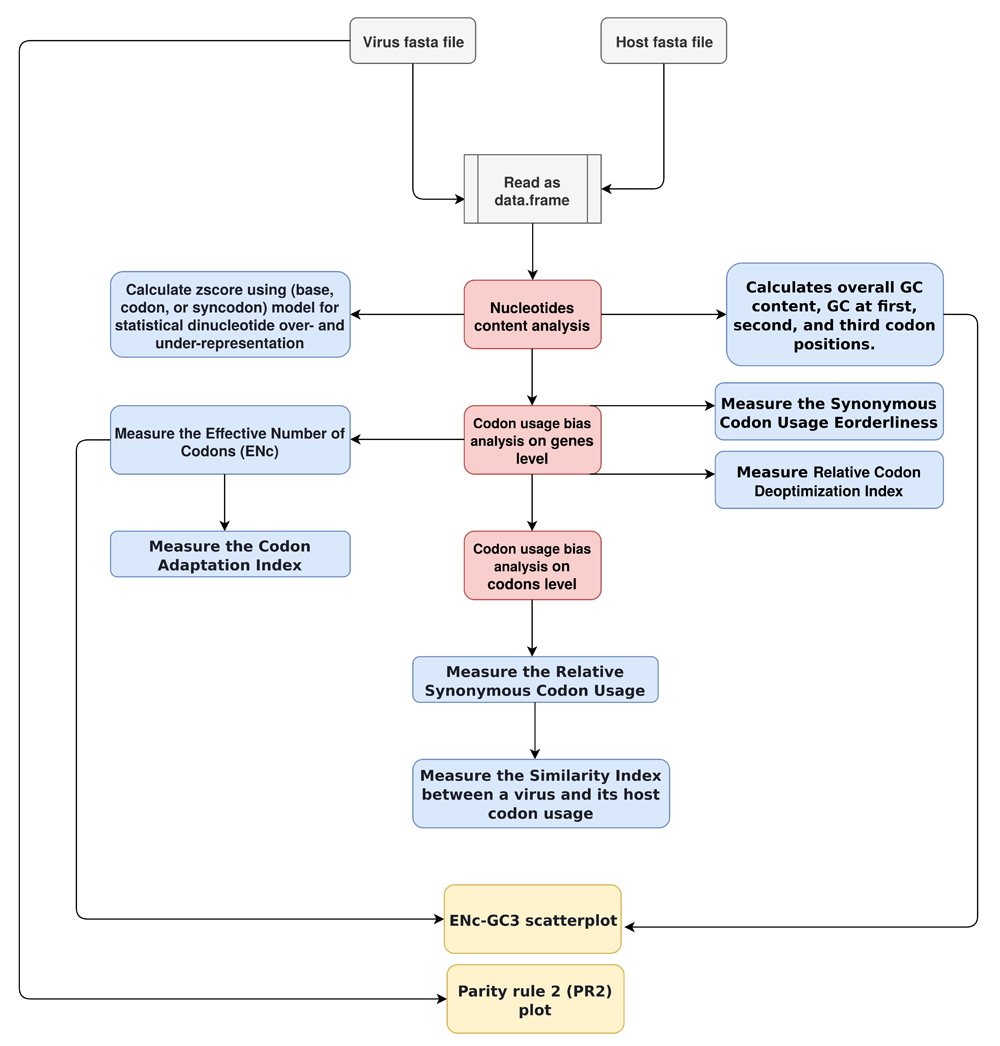
The white boxes represent the input fasta files. The red boxes represent three main analysis, each with different measures (the blue boxes), and the orange boxes represent ENc-GC3 plot and PR2-plot.

Using vhcub to study the CUB of a virus, its host and the co-adaptation between them is straightforward. As an example, we have used the coding sequences for Escherichia virus T4 and its host Escherichia coli in the form of fasta format.
# First to call the library library("vhcub") # To read both files at the same time as a data.frame # Using fasta.read() function # virus.fasta = directory path to the virus fasta file # host.fasta = directory path to the host fasta file. fasta <− fasta.read (virus.fasta = "EscherichiavirusT4.fasta", host.fasta = "Escherichiacoli.fasta") fasta.T4 <− fasta[[1]] fasta.Ecoli <− fasta[[2]]
As mentioned before, each category of analysis could be applied independently. Hence, this example will show only the codon usage bias analysis at the codon level.
# To estimate the similarity index (SiD) between E.coli T4 virus and E.coli #First Calculate the Relative Synonymous Codon Usage (RSCU) for both of them rscu.T4 <− RSCU.values(fasta.T4) rscu.Ecoli <− RSCU.values(fasta.Ecoli) # Then, the SiD could be calculated as SiD <− SiD.value(rscu.Ecoli, rscu.T4)
SiD measures the effect of the codon usage bias of the E. coli on E. coli T4 virus. In general, SiD ranged from 0 to 1 with higher values indicating that the host has a dominant effect on the usage of codons. In this example, SiD is approximately equal to 0.491. Which means that E. coli does not dominate E. coli T4 CUB. Also, this code generates RSCU values for each codon in each gene from both organisms and can be used for further analysis.
vhcub depends only on DNA sequences as input and can compute different measures of CUB for viruses, such as ENc, CAI, SCUO, and RCDI (Table 1). It can also be used to study the association between viruses and their hosts’ RSCU and SiD. There are many possible directions for future work; further versions will execute more indices, plots, and statistical analysis, to facilitate the workflow for examining the adaptations of viruses’ CUB in the R environment.
Escherichia virus T4 fasta file: ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/836/945/GCF_000836945.1_ViralProj14044
Escherichia coli fasta file: ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/005/845/GCF_000005845.2_ASM584v2/GCF_000005845.2_ASM584v2_cds_from_genomic.fna.gz
Software available from: https://CRAN.R-project.org/package=vhcub
Source code available from: https://github.com/AliYoussef96/vhcub
Archived source code as at time of publication: http://doi.org/10.5281/zenodo.357239118
License: GPL-3
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the rationale for developing the new software tool clearly explained?
Yes
Is the description of the software tool technically sound?
Yes
Are sufficient details of the code, methods and analysis (if applicable) provided to allow replication of the software development and its use by others?
Yes
Is sufficient information provided to allow interpretation of the expected output datasets and any results generated using the tool?
Partly
Are the conclusions about the tool and its performance adequately supported by the findings presented in the article?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Virology, molecular biology, infectious diseases
Is the rationale for developing the new software tool clearly explained?
Yes
Is the description of the software tool technically sound?
Yes
Are sufficient details of the code, methods and analysis (if applicable) provided to allow replication of the software development and its use by others?
Yes
Is sufficient information provided to allow interpretation of the expected output datasets and any results generated using the tool?
Yes
Are the conclusions about the tool and its performance adequately supported by the findings presented in the article?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Viral immunology
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 23 Dec 19 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)