Keywords
Canine impaction, PAX9 gene, PCR, sequencing DNA, SNPs, Genotype
Canine impaction, PAX9 gene, PCR, sequencing DNA, SNPs, Genotype
Maxillary canine teeth are the second most common targets of impaction after the third molars1. An impacted maxillary canine occurs in 1% to 3% of the general population and is twice as common in females as males2,3. It is commonly presented in clinics by patients often arriving with an aesthetic-related complaint.
Maxillary canine impaction in the palatal position is possibly caused by genetic factors and often accompanied by dental abnormalities in tooth shape, size, number, and structure. Abnormalities, such as agenesis, oligodontia, and peg-shaped teeth, have a genetic link to the presence of impacted teeth and generally manifest in developmental disorders during growth4–8. There is a relationship between malposition of certain teeth, such as palatal canines, and teeth agenesis. Similar to dental agenesis, canine-tooth-position anomalies affect several family members and are considered to be under strong genetic control9–15. Paired-box gene 9 (PAX9) is most commonly involved in affecting the odontogenesis process and thought to determine the localization of tooth seeds16. In this study, we identified an association between PAX9 genotype and the occurrence of maxillary canine impaction.
Patients were recruited from the Dental Hospital Faculty of Dentistry, Universitas Indonesia, and three different junior high schools in South Jakarta, and the study was conducted through clinical observations from May 2018 to August 2018. Those meeting the inclusion criteria (male or female, 10–25-years old, no systemic disease, and no hereditary disease) were either diagnosed with maxillary canine impaction (group I) or diagnosed as without (group II; control). Diagnosis was based on clinical examinations and radiographic interpretations performed by radiologists and orthodontists who were experts in their fields. Comprehensive clinical data were obtained for 132 patients (see Underlying data17). All participants gave their written informed consent to participate in this study, which was approved by the Ethics Committee of the Faculty of Dentistry, Universitas Indonesia (No. 07107105/Ethical Approval/ FKGUI/2018).
Genomic DNA was collected from the buccal mucosa via swabbing and extracted using the Gene Jet whole blood genomic DNA purification kit (Cat. No. K0781; Thermo-Biogen, Karlsruhe, Germany). The area of the buccal mucosa/cheek to be treated was dried with a cotton roll to prevent salivary contamination. Samples from the buccal mucosa were obtained using a cytobrush (#C0104; Medscan; Cooper Surgical, Trumbull, CT, USA) on the bilateral buccal mucosa, with each side swabbed 10 to 15 times. The cytobrush swab was inserted into a screw-capped Eppendorf tube (Cat. No. SPL-60015; Extragene, Taichung City, Taiwan) containing 200 µL of 1% phosphate-buffered saline. Eppendorf tubes were labeled and stored in a freezer at −4°C. DNA concentration was measured using a Qubit Fluorometer 3 (#Q33216; Invitrogen, Carlsbad, CA, USA) at standard fluorescence wavelengths (excitation/emission: ~480/530 nm) with Qubits assay reagent (MP423-#Q32851; Qubit dsDNA HS assay kit; Invitrogen).
We used four sets of primers spanning the PAX9-coding region (exons 2, 3, and 4) (Table 1) and standard PCR procedures for amplification of genomic DNA. PCR was performed using a T100 thermal cycler (No. #186-1096; Bio-Rad, Hercules, CA, USA) in a total volume of 25 µL containing 20 µL master mix (MyTag, 12.5 µL; Primer F, 0.5 µL; Primer R, 0.5 µL; and nuclease-free water, 6.5 µL) and 20 ng DNA. Samples were initially heated to 95°C for 10 min, followed by 30 cycles of denaturation at 95°C for 2 min, primer annealing at the optimal annealing temperature for the PAX9 primer for 1.5 min (optimal annealing temperature for PAX9 primers were as follows: F1-R1, 60°C; F2-R2, 64°C; F3-R3, 60°C; F4-R4, 56°C to 64°C), and extension/elongation at 72°C for 2 min then a final extension at 72°C for 15 min.
The PCR products were analyzed by electrophoresis using a 2% agarose gel (UltraPure Agarose; Cat. No. #16500500; Thermo Fisher Scientific, Waltham, MA, USA). To make the 2% agarose gel, 2 g agarose was added to 100 mL Tris base-acetate-EDTA (TAE) buffer, followed by the addition of 1 µL of GelRed nucleic acid gel stain (Cat. No. #41003-1; Biotium, Fremont, CA, USA). The wells were loaded with 5 µL of sample, with a DNA standard added to one well ; Cat. No. #SM0241; Thermo Fisher Scientific), and the gel was run at 100 V for 30 min.. The results of gel electrophoresis were visualized by using UV-transiluminator Gel DocTM 2000 (Cat. No. #170-8101; Bio-Rad, Hercules, CA,, USA).
DNA from the PCR products was purified using an QIAquick PCR purification kit (Cat. No 28106; Qiagen, Hilden, Germany), and purified DNA was sequenced by First-Base Laboratories (Selangor, Malaysia). Sequencing data were edited using BioEdit software (v.7.0.9; Ibis Therapeutics, Carlsbad, CA, USA) and verified using NCBI BLAST (GenBank accession No. NG_0133557.1:5001-25240).
To detect sites containing potential single nucleotide polymorphisms (SNPs), sequencing data were converted to FASTQ format, and chromatogram-peak of each patient DNA sequence was performed as can be seen in the representative data from one sample (Figure 1). Heterozygous SNPs were determined by visually identifying sites containing two overlapping peaks in both the forward and reverse sequences.
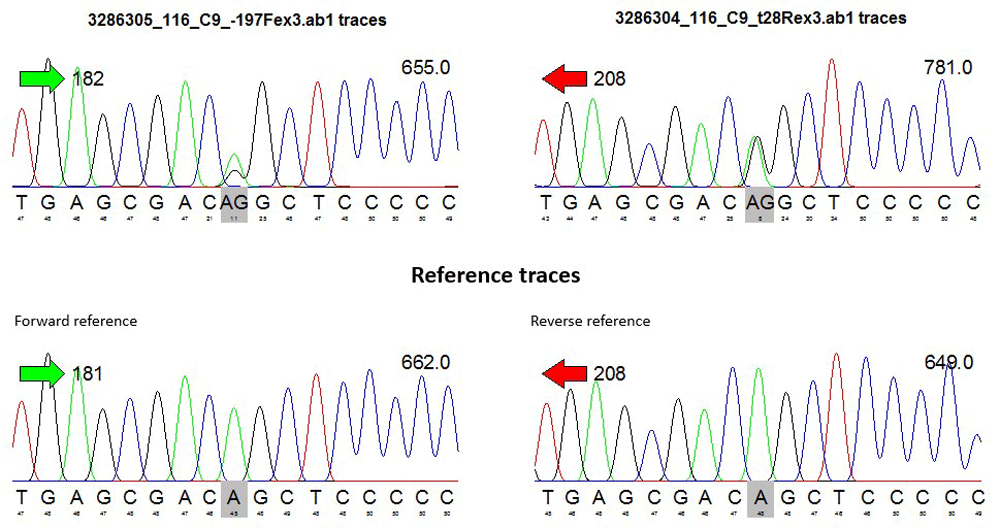
For each SNP identified, genotypes across all samples were obtained, annotated, and cross-checked against the NCBI SNP reference database (dbSNP v.150). Both genotype and allele counting were performed before conducting statistical analysis for both the case and control groups.
Statistical analysis of the data was performed using SPSS (v.20.0; SPSS, IBM Corp., Armonk, NY, USA). Analysis was initiated with a quality check of the variables. Analysis of genotype data was conducted using a chi-squared test to compare SNP frequency between patients with maxillary canine impaction and controls. A P < 0.05 was considered statistically significant.
Four SNPs were identified, with all of these located in exon 3 of PAX9 sequenced using primer pair 3 (−197Fex3 and +28Rex3). Sequencing data for this region were available for 121 of 132 samples, and no SNPs were identified in regions sequenced using the other primer pairs.
Table 2 summarizes the number of SNPs identified. Of the four identified, three were reported previously in exon 3 of PAX9 and are annotated in NCBI dbSNP (v.150; rs375436662, rs12881240, and rs4904210). The fourth SNP in exon 3 maps to chromosome 14, position 36,666,530, and has not been previously reported.
| Primer pair ID | Primer in Samples | Gene | Exon | No of SNPs Identified |
|---|---|---|---|---|
| Primer pair 1 | 110R2ex2 | PAX9 | 2 | 0 |
| 109F2ex2 | ||||
| Primer pair 2 | 357R1ex2 | PAX9 | 2 | 0 |
| -58F1ex2 | ||||
| Primer pair 3 | --197Fex3 | PAX9 | 3 | 4 |
| +28Rex3 | ||||
| Primer pair 4 | -121Fex4 | PAX9 | 4 | 0 |
| +74Rex4 |
Table 3 lists the SNPs identified in this study along with their annotation/reference identification and the location of the nucleotide substitutions. Nucleotide changes included 640A>G in SNP 1, 700C>T in SNP 2, 717C>T and 718G>C in SNP 3.
Genotype assessment was performed for all of the identified SNPs, as well as controls, focusing on counts of homozygous (wild-type) and heterozygous and homozygous (mutant) alleles (Table 4–Table 7).
| Genotype Count | Allele Count | ||||
|---|---|---|---|---|---|
| SNP 1/rs375436662 | AA | AG | GG | A | G |
| Maxillary Canine Impaction | 82 | 1 | 0 | 165 | 1 |
| Control | 38 | 0 | 0 | 76 | 0 |
| Total | 120 | 1 | 0 | 241 | 1 |
| Genotype Count | Allele Count | ||||
|---|---|---|---|---|---|
| SNP 2/rs? | CC | CT | TT | C | T |
| Maxillary Canine Impaction | 82 | 1 | 0 | 165 | 1 |
| Control | 38 | 0 | 0 | 76 | 0 |
| Total | 120 | 1 | 0 | 241 | 1 |
| Genotype Count | Allele Count | ||||
|---|---|---|---|---|---|
| SNP 3/rs12881240 | CC | CT | TT | C | T |
| Maxillary Canine Impaction | 46 | 32 | 5 | 124 | 42 |
| Control | 24 | 13 | 1 | 74 | 15 |
| Total | 80 | 45 | 6 | 198 | 57 |
| Genotype Count | Allele Count | ||||
|---|---|---|---|---|---|
| SNP 4/rs4904210 | CC | CG | GG | C | G |
| Maxillary Canine Impaction | 24 | 42 | 17 | 90 | 76 |
| Control | 9 | 20 | 9 | 38 | 38 |
| Total | 33 | 62 | 26 | 128 | 114 |
For SNPs 3 and 4, all genotype and allele variations were observed in both the case and control samples, whereas for SNPs 1 and 2, the homozygous and heterozygous mutants were present in only one of the cases (Table 6 and Table 7).
We then analyzed associations between SNPs 3 and 4, which were present in both case and control samples, and maxillary canine impaction. We verified Hardy–Weinberg equilibrium (HWE) for both SNPs in case and control samples (Table 8), with the TT genotype in SNP 3 (rs12881240) showing a higher degree of association with maxillary canine impaction than those with the CC genotype [odds ratio (OR): 2.61; 95% Confidence interval (CI): 0.29–23.61] or the CT genotype (OR: 1.28; 95% CI: 0.57–2.89), although this results was not statistically significant (P > 0.05). This result indicated a similar frequency of recessive and dominant carriers of SNP 3 between case and control samples. Furthermore, no statistical difference in allele frequency was observed between the two groups.
Individuals with the GG genotype in SNP 4 (rs4904210) were less likely to have maxillary canine impaction than those with the CC genotype (OR: 0.71; 95% CI: 0.23–2.16] and the CG genotype (OR: 0.79; 95% CI: 0.31–2.00] (Table 8). Additionally, individuals were less likely to have the G allele than the C allele (OR: 0.84; 95% CI: 0.49–1.45]. However, similar to SNP 3, these results were not statistically significant (P > 0.05).
The use of patient genotype information for clinical assessment represents a possible diagnostic strategy for predicting disease likelihood. Patient characteristics and genotype information were available from 121 of 132 samples, with Table 9 summarizing the clinical information associated with both the case and the control groups.
Stratification of cases with SNP 3 (rs12881240) showed that 21 of 53 males and 24 of 68 females in the case group harbored the CT genotype, whereas only four males and two females harbored the TT genotype, with the difference between males and females in this group not statistically significant (P = 0.3996). Additionally, the OR according to gender was 0.76 (95% CI: 0.36–1.62) for individuals harboring the CT genotype of SNP 3 (rs12881240) relative to the CC genotype, whereas it was 0.33 (95% CI: 0.06–1.94) for individuals harboring the TT genotype relative to the CC genotype.
For SNP 4 (rs4904210), 26 of 53 males and 36 of 68 females in the case group had a CG genotype, whereas 13 males and 13 females had the GG genotype. Additionally, the OR according to gender was 1.02 (95% CI: 0.43–2.40) for individuals harboring the CG genotype of SNP 4 (rs4904210) relative to the CC genotype, whereas it was 0.74 (95% CI: 0.26–2.07) for individuals harboring the GG genotype relative to the CC genotype.
Although the frequency of genotype variation was higher in females than in males, the difference was not statistically significant (P = 0.7725). Consequently, these findings suggested that gender was not a major influence on the occurrence of maxillary canine impaction.
The etiology of canine impaction might be multifactorial and involve external factors, such as environmental input (e.g., trauma), local factors (e.g., lack of space, prolonged retention of primary teeth, trauma to permanent tooth seeds, rotation of permanent seed teeth, and the presence of pathological lesions, such as dentigerous cysts or odontoma), genetic factors, and systemic disease18–24. Dentistry is increasingly making use of genetic information, which plays an important role in addressing clinical problems, especially with regard to dental abnormalities or anomalies, including impaction of the maxillary canines. Previous studies show that tooth development is regulated by >200 genes23. PAX9 encodes a transcription factor and is among the most frequently identified genes affecting the odontogenic process and involved in the occurrence of dental anomalies, such as agenesis teeth, congenital missing teeth, and variabilities in tooth size and position. The identification of genetic risk factors associated with canine impaction has recently become the subject of intensive research.
In this study, DNA sequencing of 121 of 132 patient and control samples identified four SNPs located in a similar region of PAX9 exon 3 (Table 2). SNPs play a role in determining disease characteristics, including etiology and the incidence and risk of disease development. Subsequent analysis revealed that all of the identified SNPs would result in missense mutations (Table 3). These findings suggest that SNPs might be efficacious for determining dental anomalies, specifically the impaction of maxillary canines.
Although we found no statistically significant association between PAX9 genotype and maxillary canine impaction (Table 8), there were variations between patients with and without this condition according to the presence of SNP 3 and SNP 4, which carried a greater risk for maxillary canine impaction. A previous study reported that the genes involved in the impaction or displacement of canines into the palate are also responsible for controlling the growth and eruption of teeth25, and Klein et al.26 showed that dental anomalies (size, shape, and position of teeth and agenesis of teeth or supernumerary teeth) were determined by a set of genes involved in tooth development. The results of the present study suggested a potential role for PAX9 in tooth growth and development.
In summary, our findings showed no statistically significant association between SNP genotype and gender and demonstrated that although the frequency of impaction-related genotype variation in women was higher than that in men, the differences were not statistically significant. These results suggested that gender-associated variations in genetic profile do not contribute to the incidence of maxillary canine impaction.
Written informed consent for publication of patient details was obtained.
ABI files and chromatograms that support the findings of this study are available on reasonable request from the corresponding author [author initials] and by submitting the applicable request form (‘Form Padia’ to request access for genetic resources (available as part of the OSF deposit)). The data are not publicly available due to them containing information that could compromise research participant privacy.
Open Science Framework: Genotyping Analysis Pax9 Gene In Patients With Maxillary Canine Impaction. https://doi.org/10.17605/OSF.IO/B37CJ17.
This project contains the following underlying data:
Open Science Framework: Genotyping Analysis Pax9 Gene In Patients With Maxillary Canine Impaction. https://doi.org/10.17605/OSF.IO/B37CJ17
This project contains the following extended data:
Data are available under CC0 1.0 Universal Public Domain Dedication
We are grateful for the editage for their copyediting services. We also thank you to Dr. Miesje Karmiati who helped establish the clinical diagnosis of canine tooth impaction cases in this study. Lastly, we thank Astridevitanti (Vivi) who helped work on PCR in Oral Biology Laboratory, Faculty of Dentistry UI.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Oral Biology
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Cell Biology
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 05 Mar 19 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)