Keywords
Eucalyptus sp., seed, tripsin inhibitor.
Eucalyptus sp., seed, tripsin inhibitor.
The high biodiversity of plants has led to them becoming of increasing interest to research communities due to their potential for providing new bioactive molecules with new mechanisms of action (Viegas et al., 2006). Among the most important studied plant structures, seed extracts have demonstrated potential biological activities such as protease inhibition, hemagglutinating, antibacterial and anticoagulant activities (Otieno & Analo, 2012).
The seed is the structure of a plant responsible for the propagation, and dispersion of plants in the environment, as well as nourishing and protecting the embryo at the first critical stages of germination and establishment in soil (Mello et al., 2010). To fulfill such functions these organs require a true arsenal of molecules, such as carbohydrates, lipids, amino acids and proteins (Banik et al., 2018; Mello et al., 2010).
Eucalyptus belongs to Myrtaceae family (Otieno & Analo, 2012) and some species of this genus are used in the treatment of certain bacterial or fungal infections in humans. Eucalyptus monoculture provides distinct products, such as wood, charcoal, resins, plywood, cellulosic ethanol, cellulose and paper (Takahashi et al., 2004). The present study had as objective to evaluate the protein profile and to test the hemagglutinating, hemolytic and anticoagulant activities, as well as the antitryptic effect of the crude extracts and fractions obtained from the seeds of Eucalyptus sp.
The seeds used in the present study were donated by the seed bank of the National Forest (Flona) of Nísia Floresta, located in the district of Nísia Floresta, Rio Grande do Norte, Brazil. They were powdered using refrigerated mill (TE® 631/2) until obtaining a fine flour. The ground seeds were then homogenized in 0.02 M sodium phosphate buffer, pH 6.0, under constant stirring using magnetic stirrer (Solab® SL-91/A) for 4 hours at 4°C. The homogenate was centrifuged (Hettich® MIKRO 200/200R) at 10.000 x g for 30 minutes at 4°C. The supernatant was termed crude extract (EB). EB was sequentially fractionated in two steps (0–30% named after F1 and 30–60% named as F2) with ammonium sulfate at 30% (w/v) then 60% (w/v) and further centrifuged at 10,000 x g for 30 minutes at 4°C. The pellet was resuspended in distilled water and dialyzed against its same solvent. Protein quantification was performed according to the method described by Bradford (Bradford, 1976) with adaptations for microplate assay. Plate reading was performed at 595 nm using EPOCH (Biotek®) microplate reader.
The electrophoretic protein pattern of Eucalyptus sp. fractions were observed by SDS-PAGE 12.5% (SDS-PAGE kit 1615100, Bio-Rad®) according to Laemmli (Laemmli, 1970). The protein bands also were visualized by silver staining and the approximate molecular mass were estimated by SDS-PAGE using as reference the molecular weight (Kaleidoscope™, Bio-Rad®) and migration pattern of Bovine Serum Albumin (BSA) (code A9418, Sigma-Aldrich®).
Human red blood cells (from blood bags generously donated by the Hemocentro Dalton Cunha, Rio Grande do Norte, Brazil) from different types (A, B and O) treated with papain or trypsin (both of them at 0.5 mg/mL) were incubated with serial dilutions of EB, F1 or F2 in saline solution (NaCl 0.15 M) in a 96-well plate, at a ratio of 1:1. The plate was incubated for 1 hour (at pH 7.4 and 22°C), and a negative control (saline solution and red blood cells) was performed for further comparison. The degree of agglutination was visually analyzed and the titre expressed in hemagglutination unit (U.H.), which is defined as the inverse of the highest dilution where Red Blood Cells (RBCs) agglutination was observed.
RBCs were separated from the plasma by sedimentation and washed three times with saline solution. Then, 100 μL of the red blood cell suspension were incubated with 100 μL of the samples (EB, F1 or F2) for 60 minutes at 25°C. For positive control, 100 μL of RBCs suspension was incubated with 100 μL of 1% Triton X-100, while 100 μL of saline was incubated with same volume of RBCs suspension for negative control. After incubation, the reaction mixture was centrifuged (Hettich® MIKRO 200/200R) at 3.200 x g for 5 minutes at 25°C. Aliquots of 100 μL of supernatants were transferred to 96-well plates and analyzed by spectrophotometry with readings at 405 nm (Pharmacia Biotec® Ultrospec 2100 pro). The mean and standard deviation was determined by three replicate assays.
In order to evaluate the capacity of trypsin inhibition by Eucalyptus sp. seeds protein, the test was performed according to Xavier-Filho (Xavier-Filho et al., 1989), where aliquots of 10 μL of bovine trypsin (code T8802, Sigma-Aldrich®) solution (0.3 mg/ml in 50 mM tris-HCl buffer, pH 7.5) were preincubated with 120 μL of 2.5 mM HCl as well as 320 μL of 50 mM Tris-HCl buffer, pH 7.5 and 50 μL of samples from Eucalyptus sp. for 15 minutes at 37°C. After this period, the reaction was started by adding 200 μL of 1% (w/v) Azocasein (code A 2765, Sigma-Aldrich®) solution for another 30 min. The reaction was finally stopped by adding 300 μL of 20% TCA. The reaction mixture was centrifuged for 12 min at 10.000 x g and 500 μL aliquot of the supernatant were alkalinized with 500 μL 2N NaOH. The effect of the fractions on the proteolytic activity was monitored by spectrophotometer (Pharmacia Biotec® Ultrospec 2100 pro) in the 410 nm wavelength.
Human blood was added to tubes containing sodium citrate and centrifuged at 3.200 x g for 5 minutes at room temperature for separation of plasma and red blood cells. This assay was performed with serial dilutions of the samples (EB, F1 and F2) in 0.15 M PBS buffer, pH 7.4. Aliquots of 90 μL plasma were mixed with 10 μL of samples in different protein amounts (100, 25, 12.5, 6.25, 3.12, 1.56, 0.78, 0.39 μg) and incubated for 3 minutes at 37°C. Then, 100 μL of 25 mM calcium chloride was added, after 1 hour the presence or not of coagulation was observed. For the negative control, 0.15 M PBS buffer, pH 7.4 with plasma was used. The tests were adapted from the United States Pharmacopeia (1965).
The electrophoretic profile showed two major bands, one near the region corresponding to the molecular weight of bovine serum albumin (BSA), in this case, used as a marker with a known molecular mass of 66 kDa, and a second band presenting a lower molecular weight, analyzed by linear regression for the used molecular marker as performed by Dos Santos (Dos Santos et al., 2018)(Figure 1).
EB and fractions were tested for their capacity to inhibit the activity of trypsin, a serine protease. The inhibition of trypsin was calculated based on the mass of protein estimated by the Bradford method in each sample. The results showed higher specific inhibitory activity for F1 when compared to the other fractions (Table 1). Since just a small group of proteins with same shared hydrophobicity indices are equally precipitated on each ammonium sulfate fractionation step, it increases the specific activity of proteins (de Oliveira et al., 2018) leading to a higher activity as previously described (Kunitz & Northrop, 1936; Oliveira et al., 2009).
The EB and fractions (F1 and F2) obtained from the seeds were tested for anticoagulant activity and the results obtained are presented in Table 2. Anticoagulant activity was identified in all tested samples, with the highest activity exhibited by F1 with 12.5 μg of protein. Protease inhibitors, such as those studied in the present work, have the potential to inhibit blood coagulation cascade proteases arising as potential hemostasis modulators with clinical application on the treatment of blood clots (Harish & Uppuluri, 2018; Tagnon & Soulier, 1946).
The amount of protein present per dilution is expressed in μg. (-) Absence of coagulation; (+) Presence of coagulation.
| Sample | Sample protein content (µg) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 100 | 50 | 25 | 12.50 | 6.25 | 3.12 | 1.56 | 0.78 | 0.39 | |
| EB | - | - | - | + | + | + | + | + | + |
| F1 | - | - | - | - | + | + | + | + | + |
| F2 | - | - | - | + | + | + | + | + | + |
The extract and fractions were tested with erythrocytes A, B and O, treated separately, with papain and trypsin, in triplicate (Figure 2 and Figure 3). EB showed maximum titer 512 U.H. of hemagglutination for all blood types treated with papain (code P4762, Sigma-Aldrich®), and a minimum of 64 U.H. for blood B untreated with enzymes. The F1 fraction had a maximum titer of 1024 U.H for A and O erythrocytes treated with papain and trypsin, presenting the same titer for papain-treated B erythrocytes. The lowest titer of hemagglutination obtained for F1 was 256 U.H for untreated B and O types erythrocytes as described for other studies on protein seeds fractionation (Braga et al., 2015; Vodkin & Raikhel, 1986).
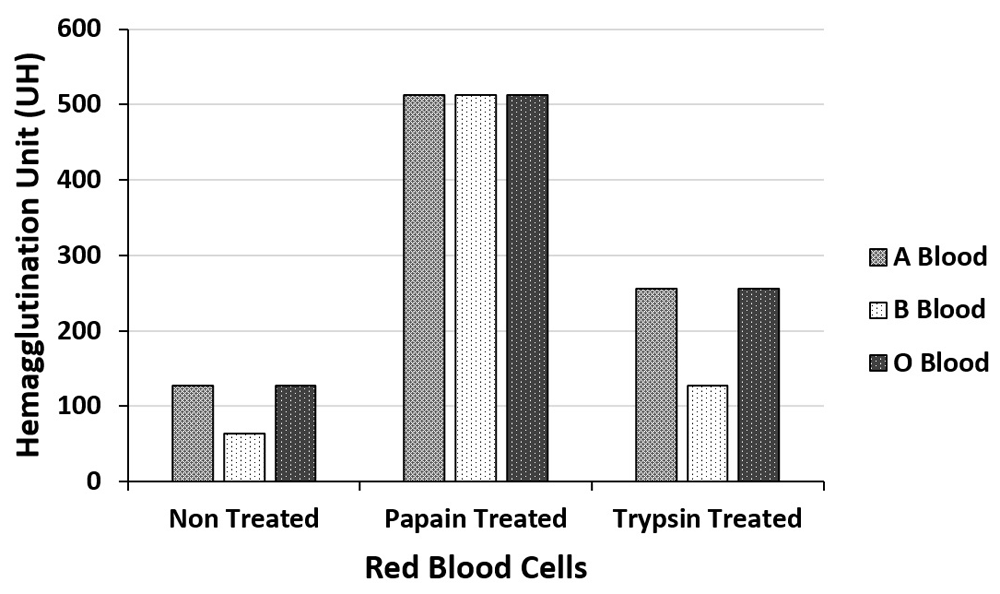
titer expressed in hemagglutination units (UH).
None of the tested samples showed hemolytic activity for any blood type even at concentrations as high as 300 μg/mL (Table 3), wich makes possible an eventual intravenous administration requiring, nonetheless, a further cytotoxicity studies in order to evaluate a proper pharmacological concentration, since same property was described for others trypsin inhibitor molecules (Sintsova et al., 2018).
Saline phosphate buffer (PBS) and 1% Triton X-100, were respectively used as negative (-) and positive (+) control for hemolytic activity.
| Sample | Type A Blood | Type B Blood | Type O Blood | PBS | Triton X-100 |
|---|---|---|---|---|---|
| EB | - | - | - | - | + |
| F1 | - | - | - | - | + |
| F2 | - | - | - | - | + |
In summary, this study described the extraction and fractionation of proteins from Eucalyptus sp. seeds with antitryptic and hemagglutinating activities, suggesting the possible occurrence of bioactive proteins like lectin and protease inhibitors. In addition, none sample showed hemolytic activity against human erythrocytes. Taken together, the obtained results demonstrate the biotechnological potential of Eucalyptus sp. seeds, being still necessary to perform further studies in order to better isolate as well as describe the bioactive compounds to be detected.
Open Science Framework: The underlying data generated in the present study, https://doi.org/10.17605/OSF.IO/KFAEX (Santos, 2018).
Data are available under the terms of the Creative Commons Zero “No rights reserved” data waiver (CC0 1.0 Public domain dedication).
This work was supported by the Federal University of Rio Grande do Norte as well as by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
Partly
If applicable, is the statistical analysis and its interpretation appropriate?
No
Are all the source data underlying the results available to ensure full reproducibility?
Partly
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Biochemistry.
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
No
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Protein chemistry, platelet biology, thrombosis and hemostasis, nanotechnology.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 08 Jan 19 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)