Keywords
Infertility, mild intellectual disability, insertion
Infertility, mild intellectual disability, insertion
Several factors have been implicated in the pathogenesis of infertility, which are associated with both men and women. Infertility may also occur due to a combined etiology where both male and female factors are combined. In approximately 40% of the cases, the etiology is unclear1.
Balanced chromosomal rearrangements (BCRs), such as autosomal reciprocal translocations, can be related to infertility. The frequency of BCRs in azoospermic men and oligozoospermic men is 0.6% and 1.7%, respectively2.
Intellectual disability (ID) is characterized by significant impairment of intellectual function, adaptive daily life skills and social skills. ID is separated into five groups: mild, moderate, severe, profound and unable to classify3.
ID occurs most likely due to a genetic etiology including BCRs. ID is thought to affect about 1% of the population, of which 85% have mild ID. People with mild ID are slower nearly in all areas of intellectual development, social and daily life skills. These individuals can take care of themselves, learn basic skills associated to safety and health and they may acquire practical life skills4.
In this study, we define an azoospermic male with mild ID who was found to have a novel insertional chromosomal abnormality on conventional cytogenetic and molecular cytogenetic techniques.
A 39-year-old man was referred to our clinic due to infertility. His height and weight were 175 cm and 82 kg, respectively. The patient left school when he was in the third grade of primary school because of learning issues. He was unable to read and write properly, and had deficits in intellectual ability like reasoning or problem solving. Currently, the patient was working as a cleaner in a factory. He was noted to have mild ID.
On physical examination, the patient had no dysmorphic features. He was married for 8 years; he and his wife were not consanguineous. His parents had two children and the family history of the patient was remarkable for a deceased brother at the age of 15 years, who had also ID (Figure 1). Since there was no history of spontaneous pregnancy during his marriage, the patient was considered to represent a case of primary infertility who had also mild ID. Sperm analysis showed complete azoospermia. In vitro fertilization was performed four times by testicular sperm extraction without success. Luteinizing hormone, follicular stimulating hormone and testosterone levels were compatible with hypergonadotropic hypogonadism (Table 1). Y micro-deletion analysis demonstrated that AZFa, AZFb and AZFc regions on the Y chromosome were intact. After conventional cytogenetic analysis, we performed array conventional cytogenetic technique (aCGH). Karyotype analysis could not be performed for the parents or the patient’s brother, since they were not alive.
| Hormone | Patient | Reference range |
|---|---|---|
| Luteinizing hormone (IU/L) | 14.87 ↑ | 1.7–8.6 |
| Testosterone (ng/dL) | 1.89 ↓ | 2.18–9.06 |
| Follicular stimulating hormone (mIU/ml) | 24.25 ↑ | 1.5–12.4 |
Conventional cytogenetic technique: Peripheral blood lymphocytes were used for a 72-hour culture. Chromosome analysis was performed on phyto-haemagglutinin-induced peripheral blood lymphocytes. Metaphase plaques were analyzed using the GTG banding method at almost 500–550 band resolution.
aCGH: An Agilent SurePrint G3 CGH+SNP Microarray Kit (4x180K) was used for genetic analysis of the patient. Microarray data were analyzed using Feature Extraction and Agilent Cytogenomics v4.0.3.12 software. Log ratios between -0.5 – 0.5 and variations with less than 5 consecutive probes were excluded. Genomic positions were based on GRCh37/hg19 Homo sapiens assembly.
Conventional cytogenetic analysis revealed that the patient had an insertional translocation: 46, XY, inv ins(18;2)(q11.2;q13q22) (Figure 2). Array CGH did not show any deletion or duplication (Figure 3).
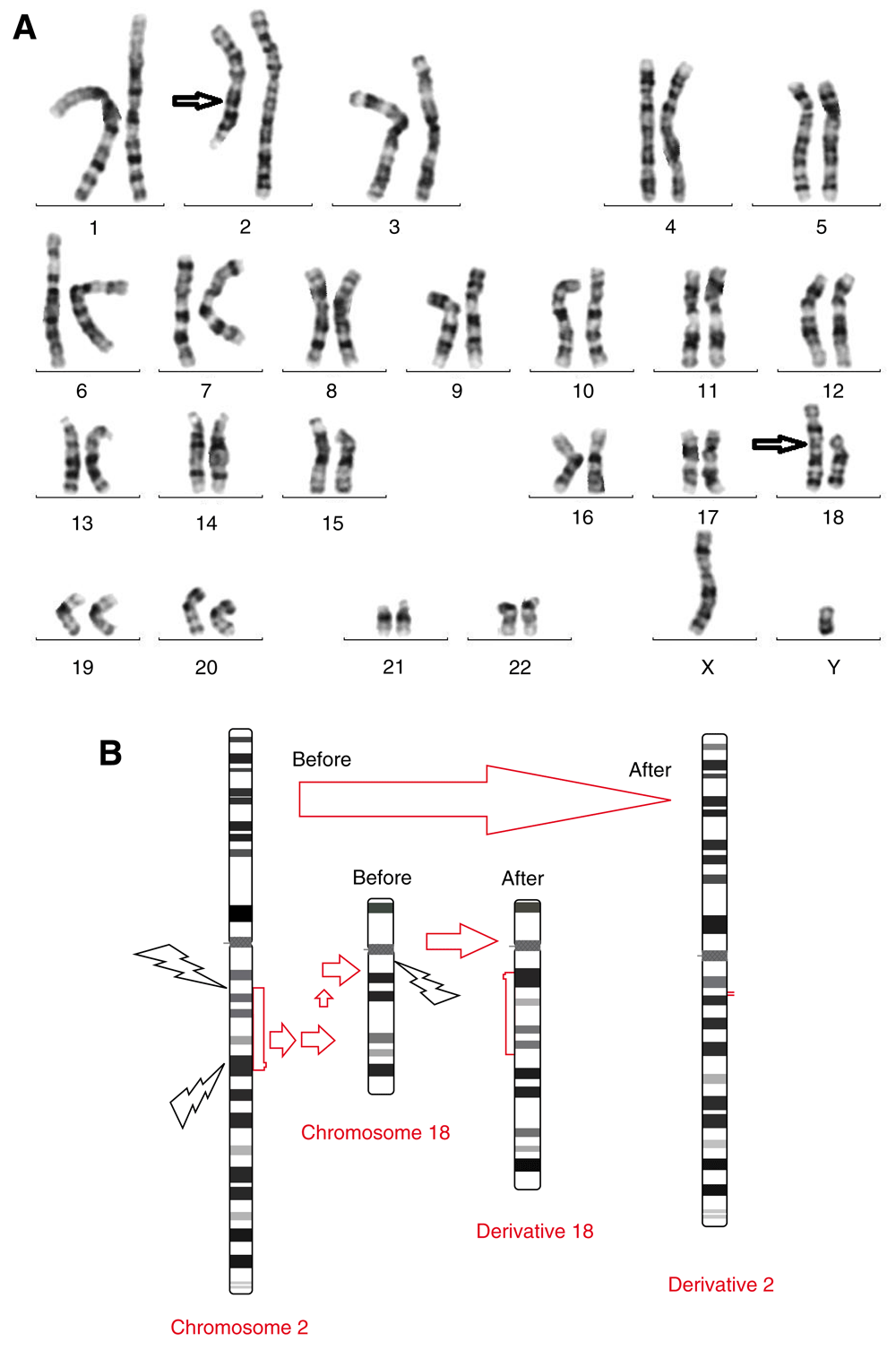
a) Patient’s GTG banded karyotype, b) Schematic of normal and derivative chromosomes 2 and 18 and breakpoints regions of chromosomes.
Insertions occur after at least three breaks on chromosomes5. Incidence of insertional chromosome translocation is nearly 1:80.000 live births6. Molecular mechanisms of chromosomal insertions are not well understood5. An individual with an inter-chromosomal insertion can transmit unblanced chromosomes to his/her child with a theoretical risk of 50% in each pregnancy6.
Insertional translocations affecting gene function may cause abnormal phenotype in different ways. According to one hypothesis, the derivative chromosomes formed after insertional translocation can contain gene or genes with increased expression. A second hypothesis states that structure of a gene located at the breakpoint may be disturbed and as a result either loss or gain of function can occur. Lastly, genes located at the breaking segment can be deleted or duplicated which can give rise to gene expression abnormalities owing to position effects6. In addition to these genetic mechanisms, insertional translocations can also affect the genome by epigenetic means. For instance, some microRNAs may be located in the break regions. It is thought that nearly 60% of all human genes are regulated by microRNAs. Furthermore, many microRNAs are located in fragile regions of the genome and considered to have an important role in the pathogenesis of many diseases7.
Classical chromosome analysis revealed that our patient carried an insertional translocation, 46, XY, inv ins(18;2)(q11.2;q13q22). To the best of our knowledge, this chromosomal insertion has not previously been reported. aCGH is used to detect genomic micro-deletions/duplications (copy number changes). However, aCGH failed to detect any micro-deletion or micro-duplication at the breakpoint regions of this insertional translocation [arr(1-22)×2,(XY)×1]. Although aCGH is a useful and a modern technique, it has some limitations. Mosaicism cannot be detected and BCR may be missed by this method. Furthermore, small deletions or duplications which are less than 10 kb can be difficult to detect with aCGH. Although conventional cytogenetic analysis is an old technique, it is still the cheapest and most useful method to obtain a general information about whole chromosomes rapidly8.
Li and colleagues elucidated the insertional translocation 46,XY inv ins(18,7) (q22.1; q36.2q21.11) found in an azoospermic man with next generation sequencing (NGS). It was demonstrated that two disrupted genes, DPP6 and CACNA2D1, were at breakpoint sites of the chromosomes, suggesting they may be associated with azoospermia. Moreover, neither micro-deletions nor duplications were detected at these breakpoint regions9.
BCRs detected by conventional karyotype analysis can be found to be unbalanced at the molecular level. We suggest that these chromosomal regions affected in our patient can be evaluated with advanced molecular techniques like NGS. Such an approach could unveil sequence abnormalities, which would potentially shed light on the molecular pathogenesis of azoospermia and/or ID.
Informed written consent for the publication of clinical details and images was obtained from the patient.
All data underlying the results are available as part of the article and no additional source data are required.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the background of the case’s history and progression described in sufficient detail?
Partly
Are enough details provided of any physical examination and diagnostic tests, treatment given and outcomes?
Yes
Is sufficient discussion included of the importance of the findings and their relevance to future understanding of disease processes, diagnosis or treatment?
Partly
Is the case presented with sufficient detail to be useful for other practitioners?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Human genetics, cytogenetics, next generation sequencing, diagnostic genetics
Is the background of the case’s history and progression described in sufficient detail?
Yes
Are enough details provided of any physical examination and diagnostic tests, treatment given and outcomes?
Yes
Is sufficient discussion included of the importance of the findings and their relevance to future understanding of disease processes, diagnosis or treatment?
Yes
Is the case presented with sufficient detail to be useful for other practitioners?
Yes
References
1. Ceylan GG, Ceylan C, Elyas H: Genetic anomalies in patients with severe oligozoospermia and azoospermia in eastern Turkey: a prospective study.Genet Mol Res. 2009; 8 (3): 915-22 PubMed Abstract | Publisher Full TextCompeting Interests: No competing interests were disclosed.
Reviewer Expertise: medical genetics
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 12 Mar 19 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)