Keywords
Transcriptomics, Bioinformatics, Software, Viral respiratory infection, Influenza viruses, Respiratory syncytial viruses (RSV), Rhinoviruses, Whole Blood, PBMC.
This article is included in the Data: Use and Reuse collection.
Transcriptomics, Bioinformatics, Software, Viral respiratory infection, Influenza viruses, Respiratory syncytial viruses (RSV), Rhinoviruses, Whole Blood, PBMC.
Viral respiratory tract infections (VRTI) are responsible for the majority hospitalizations among infants and the elderly. They are caused mainly by a heterogeneous group of viruses, including rhinoviruses, influenza viruses, parainfluenza viruses, respiratory syncytial virus (RSV), enteroviruses, coronaviruses, and certain strains of adenovirus1,2. Few antiviral therapies are currently approved and routinely used for VRTI. Most of these are specific inhibitors of influenza viruses3. Moreover, for most respiratory viruses, there is no licensed vaccine available4,5, with the exception of flu vaccines for which protection generally lasts only one flu season. Consequently, clinical management of individuals with VRTI is mostly restricted to supportive care5.
As clinical symptoms are often overlapping and are not specific for any of the viral species, it is difficult to establish a clinical diagnosis without laboratory testing1. Furthermore, clinical manifestations of VRTI are highly variable, ranging from asymptomatic infections or illness with mild symptoms (a common cold) to clinically severe disease with life-threatening complications, such as respiratory failure and in some cases may have a fatal outcome6. Infants, the elderly and patients with chronic lung or heart diseases in particular are at high risk7.
Thus, there is an evident need to better understand the molecular mechanisms underlying the disease pathogenesis, progression as well as severity of, and immunity against, VRTI among humans8. In this context, different large scale gene expression studies have been conducted using whole blood or peripheral blood mononuclear cells (PBMCs), to assess the human immune response to natural9–11 and experimental viral respiratory infections12,13; in particular, to influenza and RSV infections, and also to vaccination14–17.
Here, we make available, through an interactive web application, a curated collection of datasets that were obtained from pediatric and adult patients with natural VRTI, volunteers with experimental exposition to respiratory viruses and also vaccinated volunteers. Transcriptomics datasets were obtained from whole blood and PBMCs.
A total of 31 datasets were retrieved and selected from the NCBI Gene Expression Omnibus (GEO), a public repository of transcriptome profiles. The identified datasets are particularly relevant to our interest in understanding the pathobiology of VRTI and vaccination. As described in recent publications18,19, these datasets were loaded into a custom interactive web application, the Gene Expression Browser (GXB), which enables easy access to large datasets and interactive visualization of our dataset collection related to VRTI and vaccination against respiratory viruses. It also provides access to demographic and clinical information. Importantly, the user can customize data plots by adding multiple layers of parameters (e.g. age, gender, sample type, type of infection, type of vaccine, sample collection time), modify the sample ordering and genes, and generate links (mini URL) that can be shared via e-mail or used in publications. Therefore, we are providing here a resource enabling browsing of datasets relevant to blood transcriptional responses to VRTI and vaccination that offers a unique opportunity to identify host genes and their regulation that may be of diagnostic and/or prognostic value, or that may be tested as novel correlates of protection in subsequent studies. For example, a comparative approach of the transcriptional response signatures between experimentally infected and vaccinated individuals could be used to identify common mechanisms that define the poor health outcomes versus strong protection. The ability to pool, compare and analyze the immune responses to different infections and vaccines, in different individuals and at various age, offers a unique opportunity for a better understanding of the pathophysiology of VRTI.
A total of 120 datasets, potentially relevant to human immune responses to VRTI and vaccination, were identified in GEO using the following search query:
Homo sapiens[Organism] AND ((“respiratory syncytial virus”[DESC] OR RSV[DESC] OR metapneumovirus[DESC] OR hMPV[DESC] OR influenza[DESC] OR parainfluenza[DESC] OR rhinovirus[DESC] OR rhinoviruses[DESC] OR adenovirus[DESC] OR adenoviruses[DESC] OR HAdV[DESC] OR coronaviruses[DESC] OR HCoV[DESC]) OR (vaccine OR vaccines OR vaccination) AND (blood[DESC] OR PBMC[DESC] OR PBMCs[DESC] OR lymphocyte[DESC] OR lymphocytes[DESC] OR “B cell”[DESC] OR “B cells”[DESC] OR “plasma cells”[DESC] OR “T cell”[DESC] OR “T cells”[DESC] OR Treg[DESC] OR Tregs[DESC] OR monocyte[DESC] OR monocytes[DESC] OR dendritic[DESC] OR DC[DESC] OR DCs[DESC] OR "natural killer"[DESC] OR NK[DESC] OR NKT[DESC] OR neutrophil[DESC] OR neutrophils[DESC]) AND (“Expression profiling by array”[gdsType] OR “Expression profiling by high throughput sequencing”[gdsType]).
Most of retrieved datasets were generated from human blood and human PBMC, using Illumina or Affymetrix commercial platforms or RNA-sequencing. All the entries that were returned with this query were manually curated. The process involved reading all the descriptions available of the datasets, the study design and the GEO-linked article in pubmed. Finally, only studies using human whole blood and human PBMCs, associated with natural or experimental VRTI, or vaccination against VRTI, were retained for our dataset collection. For the retained datasets, if the platform used to generate the transcriptome profiles was not supported by GXB or if from an in vitro study, they were exlcluded from our dataset collection. Based on these criteria, 31 datasets were retained. These include datasets that were generated from whole blood or PBMCs of individuals who were either naturally (12) or experimentally infected (3) (with influenza viruses, RSV, Rhinovirus, Rotavirus) as well as from healthy, uninfected (age-matched) volunteers. The remaining 16 datasets were generated from whole blood or PBMCs of individuals who had received flu vaccines (Figure 1). The datasets that comprise our collection are listed in Table 1.
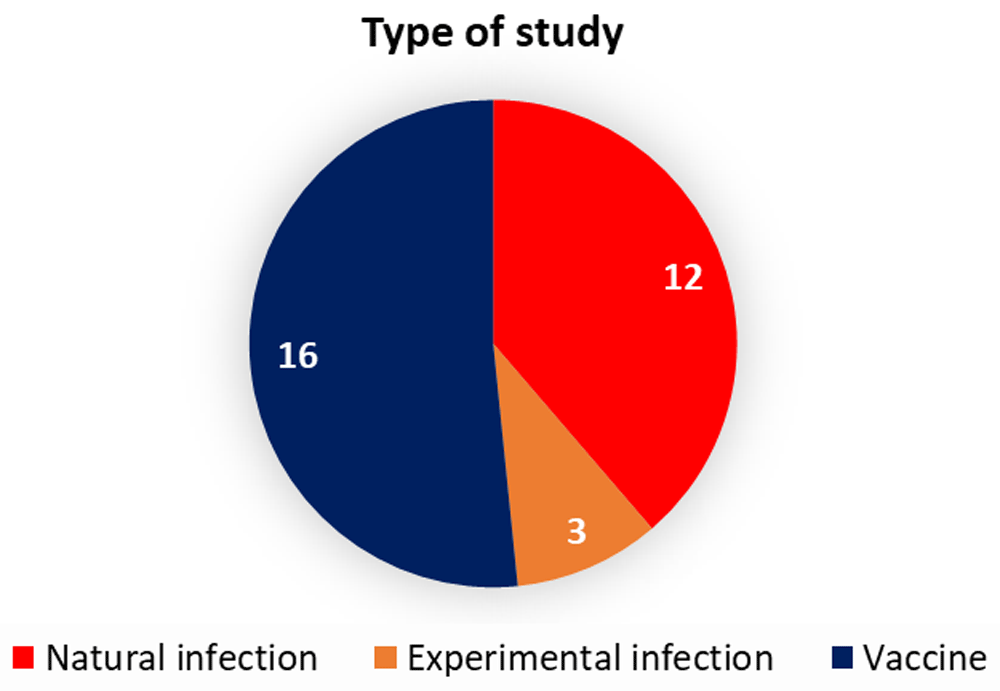
The pie chart indicates the type of studies carried out for the 31 datasets.
Once the final selection had been made, each dataset was downloaded from GEO by using the SOFT file format. Then, the datasets were uploaded on the Gene Expression Browser (GXB), an interactive web application hosted on the Amazon Web Services cloud20. Information about samples and study design were also uploaded. The available samples were put into groups based on relevant study variables and genes were ranked according to the different groups comparisons. A detailed description of the GXB software tool is available from recent publications19–21. This software interface allows user to easily navigate and filter the dataset collection. A web tutorial can be easily accessed online. Annotation and functionality of the web software interface were described previously by our group18,19,21, and is reproduced here so that readers can use this article as a standalone resource. Briefly, datasets of interest can be quickly identified either by filtering on criteria from pre-defined sections on the left or by entering a query term in the search box at the top of the dataset navigation page. Clicking on one of the studies listed in the dataset navigation page opens a viewer designed to provide interactive browsing and graphic representations of large-scale data in an interpretable format. This interface is designed to present ranked gene lists and display expression results graphically in a context-rich environment. Selecting a gene from the rank ordered list on the left of the data-viewing interface will display its expression values graphically in the screen’s central panel. Directly above the graphical display drop down menus give users the ability: a) To change how the gene list is ranked - this allows the user to change the method used to rank the genes, or to only include genes that are selected for specific biological interest; b) To change sample grouping (Group Set button) - in some datasets, a user can switch between groups based on cell type to groups based on disease type, for example; c) To sort individual samples within a group based on associated categorical or continuous variables (e.g. gender or age); d) To toggle between the bar chart view and a box plot view, with expression values represented as a single point for each sample. Samples are split into the same groups whether displayed as a bar chart or box plot; e) To provide a color legend for the sample groups; f) To select categorical information that is to be overlaid at the bottom of the graph - for example, the user can display gender or smoking status in this manner; g) To provide a color legend for the categorical information overlaid at the bottom of the graph; h) To download the graph as a portable network graphics (png) image. Measurements have no intrinsic utility in absence of contextual information. It is this contextual information that makes the results of a study or experiment interpretable. It is therefore important to capture, integrate and display information that will give users the ability to interpret data and gain new insights from it. We have organized this information under different tabs directly above the graphical display. The tabs can be hidden to make more room for displaying the data plots, or revealed by clicking on the blue “show info panel” button on the top right corner of the display. Information about the gene selected from the list on the left side of the display is available under the “Gene” tab. Information about the study is available under the “Study” tab. Rolling the mouse cursor over a bar chart feature while displaying the “Sample” tab lists any clinical, demographic, or laboratory information available for the selected sample. Finally, the “Downloads” tab allows advanced users to retrieve the original dataset for analysis outside this tool. It also provides all available sample annotation data for use alongside the expression data in third party analysis software. Other functionalities are provided under the “Tools” drop-down menu located in the top right corner of the user interface. Some of the notable functionalities available through this menu include: a) Annotations, which provides access to all the ancillary information about the study, samples and dataset organized across different tabs; b) Cross-project view; which provides the ability for a given gene to browse through all available studies; c) Copy link, which generates a mini-URL encapsulating information about the display settings in use and that can be saved and shared with others (clicking on the envelope icon on the toolbar inserts the URL in an email message via the local email client); d) Chart options; which gives user the option to customize chart labels.
Quality control checks can be performed on the datasets loaded on GXB, for example by examining concordance of the gender-specific expression of the XIST gene in those datasets for which gender information was available as metadata. The XIST gene is essential for imprinted and random X-chromosome inactivation22 and therefore, expression is expected to be high in female and low in male samples. Respective hyperlinks are found in Table 1 allow you to visualize the XIST experession based on the gender information provided with the GEO submission. Figure 2 shows XIST gene expression in a representative dataset, along with gender information available that was recorded and made available in GEO.

Gene expression data were from whole blood of healthy adult volunteers before and after receiving either placebo (saline) injections, seasonal influenza (Fluzone) or pneumococcal (Pneumovax) vaccination.
All datasets included in our curated collection are also available publically via the NCBI GEO website : https://www.ncbi.nlm.nih.gov/gds/; and are referenced throughout the manuscript by their GEO accession numbers (e.g. GSE17763). Signal files and sample description files can also be downloaded from the GXB tool under the “downloads” tab.
All the authors listed on this publication received support from the Qatar Foundation. Support for this project was provided by the Qatar National Research Fund [NPRP10-0205-170348].
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We would like to thank all the investigators who decided to make their datasets publically available by depositing them in GEO.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the rationale for creating the dataset(s) clearly described?
Yes
Are the protocols appropriate and is the work technically sound?
Yes
Are sufficient details of methods and materials provided to allow replication by others?
Yes
Are the datasets clearly presented in a useable and accessible format?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Genomics, immunology, viral respiratory infection
Is the rationale for creating the dataset(s) clearly described?
Yes
Are the protocols appropriate and is the work technically sound?
Yes
Are sufficient details of methods and materials provided to allow replication by others?
Yes
Are the datasets clearly presented in a useable and accessible format?
Yes
References
1. Speake C, Presnell S, Domico K, Zeitner B, et al.: An interactive web application for the dissemination of human systems immunology data.J Transl Med. 2015; 13: 196 PubMed Abstract | Publisher Full TextCompeting Interests: No competing interests were disclosed.
Reviewer Expertise: genomics
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 13 Mar 19 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)