Keywords
Rheumatoid arthritis, CCL28 inhibition, Molecular docking, Astragalin, Binding affinity
This article is included in the Cheminformatics gateway.
Rheumatoid arthritis, CCL28 inhibition, Molecular docking, Astragalin, Binding affinity
Rheumatoid arthritis (RA) is an autoimmune inflammatory disease of synovial joints1. The prevalence of RA is about 1% worldwide and women are three times more likely to be affected than men. The onset of RA is mostly seen in people aged 30–50 but can occur at any age2,3. In the past two decades, research has advanced understanding of RA pathogenesis which has revolutionized RA treatment and led to the development of new therapeutic agents4. Non-steroidal anti-inflammatory drugs (NSAIDs) and disease-modifying anti-rheumatic drugs (DMARDs) are groups of drugs commonly used to control and alter the progression of RA4,5. However, these medications cannot completely cure RA, are expensive and have some notable side effects. Therefore, the development of new therapeutics for RA with fewer side effects is required.
Astragalin (Kaempferol 3-O-beta-D-glucoside) is a flavonoid found in black tea, alcoholic beverages such as red wine and Cuscuta plants. Preliminary studies have shown it has potential pharmacological characteristics such as anti-inflammatory and antioxidant properties6,7. Computational analysis is an important technique for determining the potential of astragalin in inhibiting C-C motif chemokine ligand 28 (CCL28). Virtual screening tools provide an easy way to identify therapeutic targets suitable for multiple ligands8.
CCL28, a mucosa-associated epithelial chemokine, is found in the epithelial cells of many tissues such as the lungs, guts and salivary glands. Studies have reported the involvement of CCL28 in RA angiogenesis, a process that occurs early in RA pathogenesis9. RA angiogenesis is fostered by the interaction of pro-inflammatory cytokines and chemokines. The ligation of CCL28 to its receptor CCR10 is involved in B- and T-cell trafficking in RA pathogenesis10.
The ability of astragalin to bind and inhibit CCL28 has not yet been reported. Therefore, in this paper we will describe the potential inhibition of the CCL28 target by an astragalin ligand using computational and bioinformatics tools.
The three-dimensional crystal structure of the target protein CCL28 (PDB ID: 6CWS)11 was downloaded from the Protein Data Bank12 in PDB format using a similar method to that carried out by Esther et al.13
The two-dimensional chemical structure of astragalin was accessed from the PubChem database (PubChem CID: 5282102) and drawn using Accelrys Draw 4.1 software14 with default parameters. The resulting MOL file of the ligand was converted into PDB format by Accelrys Discovery Studio 4.1 software15 using default parameters. The chemical structure of astragalin is shown in Figure 1. We used naproxen as standard drug which is being used against arthritis. The chemical structure of this molecule was taken from the PubChem database (PubChem CID: 156391).
PDB coordinates of target and ligand proteins were optimized by energy minimization, 3D protonation and stable conformation parameters of Molecular Operating Environment (MOE) software16 (version 2014.0901) and files were saved in MDB format.
The quality of the CCL28 predicted protein structure was estimated using the Phyre2 (Protein Homology/analogY Recognition Engine, version 2.0) online web server17. The ProQ2 quality assessment and Ramachandran plot was applied for identifying the structural conformation of the proteins using the default parameters of the tool.
We computed and analysed the binding sites of target protein using the default parameters of the MOE software16.
A computational ligand-target docking approach was used to determine structural complexes of CCL28 with astragalin in order to understand the structural basis of these protein targets specificity. Finally, docking was carried out by MOE based on RMSD and E-score. The energy of the interaction of these derivatives with the protein targets is assigned “grid point”. At each step of the simulation, the energy of the interaction of ligand and protein was evaluated using atomic affinity potentials computed on a grid. Molecular docking was performed to analyse the binding of the astragalin ligand with the active sites of the CCL28 target protein. The optimized ligand was docked with the target using MOE software16 with default parameters as described by Esther et al.13
The quality of the CCL28 predicted protein structure was estimated by the Phyre2 web server17. Phyre2 provided a 3D structure of the target protein and the dimensions of the 3D structure in Å were X=48.506, Y=53.506 and Z=38.552. Analysis of the secondary structure of the target protein provided by Phyre2 revealed high confidence in the predicted structure, indicated by red regions in Figure 2, suggesting that the protein structure quality is high. The sequence profile graph shows that M residues are highly favourable, L and V residues are moderately favourable and remaining residues are less favourable. The ProQ2 quality assessment showed the quality of protein target and could be considered as "good" model indicating less mutation sites in the protein structure (Figure 3). The Ramachandran plot shows that approximately 98% amino acid residues of the target protein model exist in the most favoured region whereas <2% exist in the disallowed region. This Ramachandran plot representing the backbone conformation of CCL28 amino acid residues is shown in Figure 4.

The sequence alignment of protein molecule shows high confidence, indicated by red colour, while blue indicates low confidence. 49% of the structure is in alpha helices, 11% is in beta-strands and 13% is in transmembrane helices.
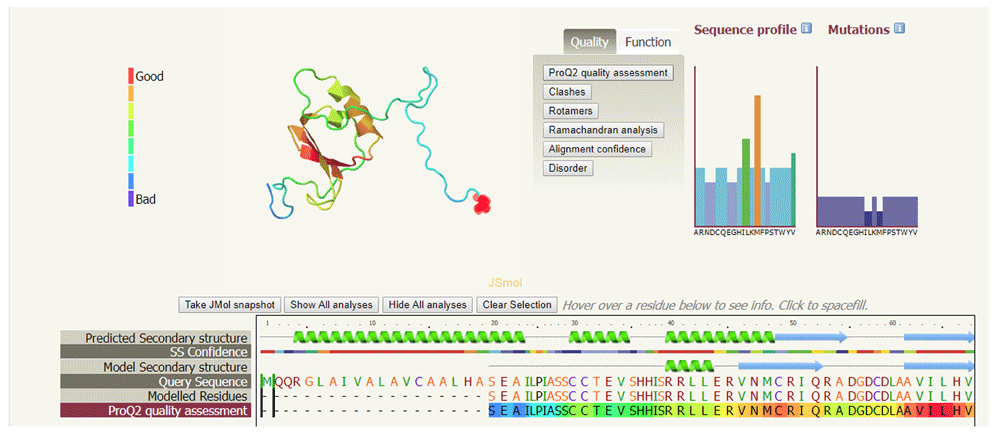
Red colour indicates “Good” regions while blue represents “Bad” regions of the structure. The quality of model indicated the minimal disordered region (shown by broken lines).
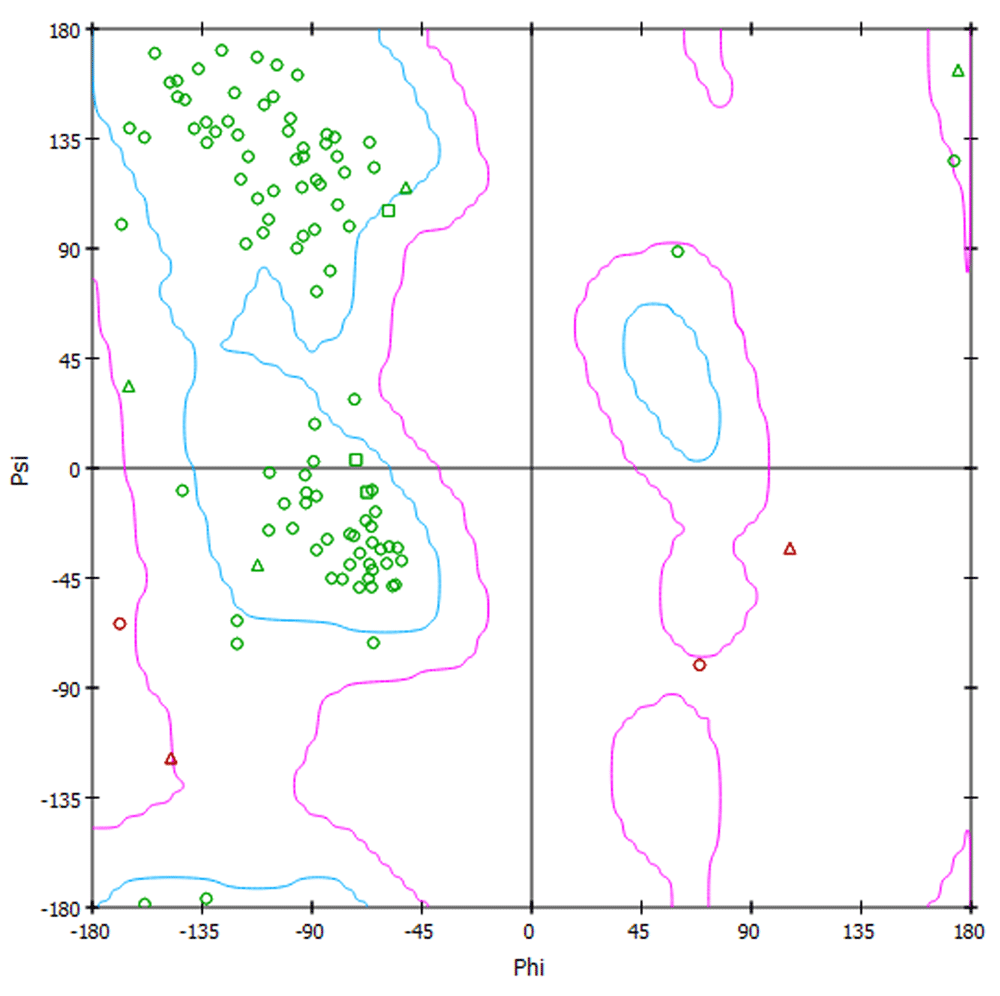
For a “good” protein model there must be ≥90% amino acid residues in the most favoured region or <2% in the disallowed region of the plot.
Binding sites of the target protein were analysed using MOE software16. Analysis revealed that His 99 and Thr 101 residues were present in a pocket of the target protein. The spatial orientation of the astragalin ligand in the binding pocket of the target is shown in Figure 5. In silico molecular docking using MOE software16 resulted in the successful docking of target CCL28 with the astragalin ligand. The binding energy of astragalin with CCL28 is -5.40 kcal/mol, as compared to standard naproxen which has a binding energy of -4.87 kcal/mol. A comparison of the binding energies and root-mean-square deviation (RMSD) values of astragalin and standard naproxen with the CCL28 target is shown in Table 1 and Table 2 respectively. Results show that astragalin has strong binding affinity for the CCL28 target molecule as compared to naproxen. The ligand interaction module of MOE16 was used to view ligand interactions. Figure 6 shows astragalin interacting with CCL28 via hydrogen bonding with the His 99 and Thr 101 residues.
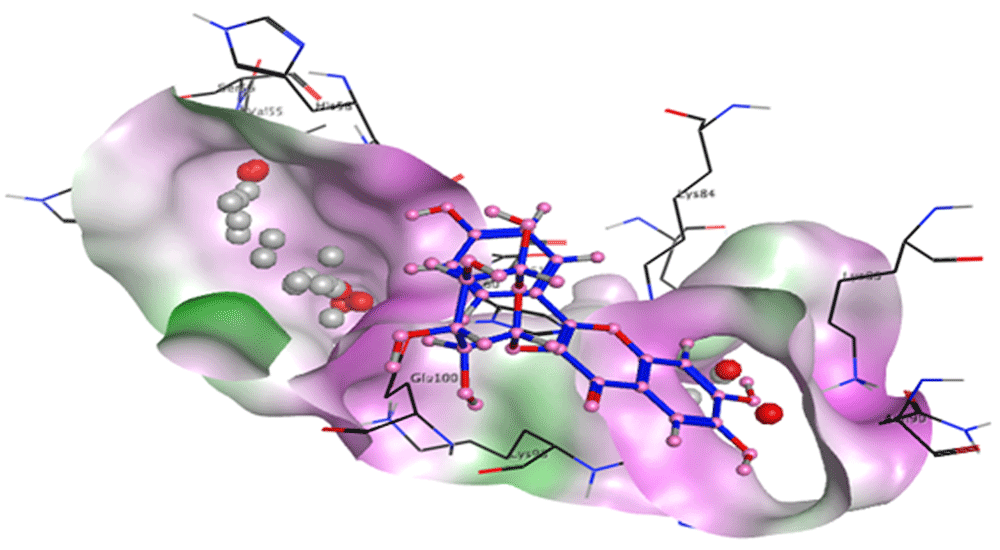
The CCL28 protein is involved in the angiogenesis of rheumatoid arthritis as it recruits leukocytes to the site of joint inflammation. Inhibition of the CCL28 protein blocks the migration of leukocytes and other proinflammatory factors and this serves as a promising target for arthritis18.
Methotrexate and leflunomide are DMARDs used for rheumatoid arthritis treatment. In a few patients, serious liver damage due to methotrexate has been observed. Similarly, leflunomide also exhibits certain side effects19. These side effects have increased the need for the development of safe alternative, treatment approaches. Current research is focused on the development of bioactive molecules from natural resources which don’t have side effects20. Astragalin isolated from the leaves of black and green tea possesses potential therapeutic effects including anti-inflammatory, antioxidant and anticarcinogenic activities. It may have the ability to inhibit the production of inflammatory mediators such as IL-1b, IL-6 and TNF-alpha19.
Astragalin acts as an inhibitor and prevents the binding of CCL28 to its receptor CCR10 which is a primary step for the recruitment of leukocytes and formation of blood vessels inside inflamed joints of RA patients21.
In silico docking studies of astragalin with CCL28 were carried out using MOE software16. Lowest binding energy and root mean square values (RMSD) from docking studies indicate a strong interaction between ligand and target. In silico molecular docking showed successful binding of CCL28 with the astragalin ligand. The binding energy of astragalin with CCL28 is -5.40 kcal/mol, a lower binding energy than that of naproxen with CCL28 which is 4.87 kcal/mol. These docking results suggest that astragalin can act as a potential inhibitor for CCL28.
Molecular docking results of astragalin with CCL28 showed that this ligand has strong binding affinity for CCL28. Therefore, astragalin may act as an important inhibitor of CCL28 to treat rheumatoid arthritis in the future.
Structure of CCL28 from Protein Data Bank, Accession number 6CWS: https://identifiers.org/pdb/6CWS.
Structure of naproxen from PubChem Database, Accession number 156391: https://identifiers.org/pubchem.compound/156391
Structure of astragalin from PubChem Database, Accession number 5282102: https://identifiers.org/pubchem.compound/5282102
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Partly
If applicable, is the statistical analysis and its interpretation appropriate?
Not applicable
Are all the source data underlying the results available to ensure full reproducibility?
Partly
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Proteomics, immune proteomics, bioinformatics, metabolomics, arthritis, Coronary artery disease
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Not applicable
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Pharmaceutical Sciences, Biochemistry.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 20 Mar 19 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)