Keywords
HSV-1, envelopment, nuclear membranes, Golgi membranes, gK, immunogoldlabeling, immunofluorescence, TEM
HSV-1, envelopment, nuclear membranes, Golgi membranes, gK, immunogoldlabeling, immunofluorescence, TEM
Capsids of herpes simplex virus 1 (HSV-1) are assembled in host cell nuclei, transported to the nuclear periphery and translocated to the perinuclear space (PNS) or into the cytosol (Roizman et al., 2007). The HSV-1 Us3 kinase phosphorylates numerous viral and cellular substrates (Kato & Kawaguchi, 2018). It is involved in nucleus-to-cytoplasm capsid translocation (Kato et al., 2009; Mou et al., 2009; Reynolds et al., 2002; Ryckman & Roller, 2004; Wisner et al., 2009), in regulation of phospholipid synthesis induced by HSV-1 (Sutter et al., 2012; Wild et al., 2012), and in blocking apoptosis induced by HSV-1 (Galvan & Roizman, 1998; Goodkin et al., 2004; Leopardi et al., 1997). The Golgi complex plays a significant role in apoptosis (Cheng et al., 2010; Hicks & Machamer, 2005), in membrane biosynthesis together with the endoplasmic reticulum (Smirle et al., 2013), and in HSV-1 envelopment (Roizman et al., 2014). Glycoprotein K (gK) is a hydrophobic transmembrane protein of the viral envelope (Mo & Holland, 1997) encoded by the UL53 gene (Bond & Person, 1984; Hutchinson et al., 1992). gK is essential for efficient envelopment by Golgi membranes (Baines et al., 1991; Melancon et al., 2007) and for production of infectious progeny virus (Chouljenko et al., 2012). HSV-1ΔUs3 virions accumulate in the PNS. Nuclear and Golgi membranes become severely altered by insertion of proteins of unknown nature (Wild et al., 2015). The essentiality of gK in envelopment by Golgi membranes prompted us to identify gK in cells infected with HSV-1ΔUs3. Because of the hydrophobicity of gK, we constructed a HSV-1ΔUs3 with a hemagglutinin (HA) tag at gK, CK177ΔUs3gK-HA. Immunogold labeling shows that gK localizes on amorphous protein structures, nuclear and Golgi membranes, and, importantly, on virions in the PNS.
Vero cells (European Collection of Cell Cultures, ECACC, 84113001) were grown in Dulbecco’s modified minimal essential medium (DMEM; 31885-023; Gibco, Bethesda, MD, USA) supplemented with penicillin (100 U/ml), streptomycin (100 μg/ml) (Anti-Anti, 15240-062, Gibco) and 10% fetal bovine serum (FBS; 2-01F10-I, Bio Concept, Allschwil, Switzerland). The Us3-deletion mutant R7041ΔUs3 was kindly provided by Bernard Roizman (The Marjorie B. Kovler Viral Oncology Laboratories, University of Chicago, Illinois, USA). Wild-type (wtHSV-1) strain F (Ejercito et al., J. Gen. Virol. 2:357–364, 1968), R7041ΔUs3 were propagated in Vero cells.
Recombineering of pYEbac102 (Tanaka et al., 2003) was done in two steps in Escherichia coli strain SW102 (Warming et al., 2005) First, a galK expression cassette was amplified by PCR (Phusion R, High-Fidelity DNA Polymerase; M0530L, BioLabs. Ipswich, MA, USA, according to manufacturer’s recommendations) with homology arms flanking UL53 using the following primers: primer forward, 5’- tct tcg gtg cca gtc cgc tgc acc gat gta ttt acg cgg tac gcc cca ccg CCT GTT GAC AAT TAA TCA TCG GCA -3’; primer reverse, 5’- gtt tcc aat ttg cat atg ccg tta cgg ttt ccg ccg gcc tgg atg tga cgt TCA GCA CTG TCC TGC TCC TT -3’ (binding sequences in capitals). This amplimer was DpnI treated to remove template DNA, purified, and electroporated into competent and induced E. coli strain SW102 carrying pYEbac102 (Tanaka et al., 2003). Recombinant colonies were selected for growth on galactose plates. The BAC DNA carrying the HSV genomic sequence with deleted UL53 was designated pYEbac102ΔUL53. Second, a kanamycin resistant cassette was amplified by PCR on pBSrspL (Genes Bridges, Dresden, Germany; kanamycin resistant cassette) with the following primers for Us3rpsL: primer forward, 5’-ctt ccc aca cca cac cac cca gcg agg ccg agc gcc tgt gtc atc tgc aGG GCC TGG TGA TGA TGG CGG GAT CG-3’, primer reverse 5’-aga tca cca gac cgg cgc tcc aaa tgt cga cgg tcg tgg tat acg gat ccT CAG AAG AAC TCG TCA AGA AGG-3’ (binding sequences in capitals). The primers were chosen to yield the same deletion of Us3 as described by Purves et al. (1987). This amplimer was DpnI-treated, purified, and electroporated into competent and induced E. coli strain SW102 carrying pYEbac102ΔUL53. Recombinant colonies were selected for growth on kanamycin plates. Finally, fHSVgKgalKΔUS3, pCS177 (HA tagged gK and flanking sequences, see below) and pCMV-Cre (Cre recombinase under CMV promoter) DNA was mixed and co-transfected into Vero cells using Lipofectamine 2000 (11668027, Thermo Fisher Scientific, Rockford, IL, USA) according to the manufacturer’s recommendations. Rescued progeny virus with deleted Us3 and BAC backbone but HA-tagged gK, designated CK177ΔUs3gK-HA, was propagated in Vero cells.
Plasmid pCS177 containing the UL53 gene with a carboxy terminal HA tag and flanking sequence to UL53 that extend 0.3 kbp upstream and 0.4 kbp downstream of the UL53 gene was constructed as follows. First, downstream sequence of UL53 was amplified by PCR from wtHSV-1 genome (primer forward, 5’- gat ctc tag acg tca cat cca ggc cgg cgg aa - 3’; primer reverse, 5’- gat cga gct cag gCC TCC GGC ACA GAC AAG GAC CAA T -3’; HSV-1 sequences in capitals). The resulting PCR product was digested with SacI and XbaI and cloned into the SacI and XbaI sites in pBluescript II KS(+), resulting in plasmid pCS176. Second, the UL53 gene with its upstream flanking sequence was amplified by PCR from wtHSV-1 using a reverse primer containing nucleotides of HA tag (primer forward, 5’- gat caa gct tag gcc tgg gtc ggt aca acg tac agc cgg at - 3’; primer reverse, 5’- gat ctc tag aTC Acc atg gag cat aat ctg gaa cat cat atg gat aTA CAT CAA ACA GGC GCC TCT gga -3’; HSV-1 sequences in capitals). Following PCR, the DNA product was digested with HindIII and XbaI and inserted into these sites in pCS176, resulting in plasmid pCS177. The StuI fragment of pCS177 containing UL53-HA gene with flanking sequence was co-transfected together with pYEbac102ΔUL53 and pCMV-Cre into Vero cells. gK-HA expression was identified using indirect immunofluorescence and one virus stock expressing gK-HA was designated CK177gK-HA.
Vero cells were grown for 2 days on cover slips (Assistent, Sondheim, Germany) for immunofluorescence, on sapphire disks (100.00174, Bruegger, Minusion, Switzerland) placed in 6 well plates for TEM, or in 75 cm2 cell culture flasks for immunogoldlabeling, for 2 days prior to inoculation with CK177ΔUs3gK-HA, R7041ΔUs3, CK177gK-HA or wtHSV-1 at a multiplicity of infection (MOI) of 1 plaque-forming unit (pfu)/ml.
Cells on sapphire disks were frozen 16 to 24 hpi in a high-pressure freezer (HPM010; BAL-TEC Inc., Balzers, Liechtenstein) and prepared for sectioning as described in detail previously (Wild et al., 2018; Wild et al., 2002). Cells were analyzed in a transmission electron microscope (CM12; FEI, Eindhoven, The Netherlands) equipped with a CCD camera (Ultrascan 1000; Gatan, Pleasanton, CA, USA).
Cells inoculated with CK177ΔUs3gK-HA or CK177gK-HA at MOI 1 were incubated for 20 h, briefly washed with PBS, fixed with 2% formaldehyde for 25 min at room temperature, washed with cold PBS, permeabilized with 0.1% Triton-X-100 at room temperature for 7 min, and blocked with 3% bovine serum albumin in phosphate-buffered saline (PBS) containing 0.05% Tween 20 (PBST). Cells were labeled with mouse monoclonal HA-probe antibodies (1:500) (SC-7392, Santa Cruz Biotechnology, Dallas, TX, USA), followed by anti-mouse Alexa 488-conjugated secondary antibodies (1:500) (A1101; Thermo Fisher) as well as with polyclonal antibodies (1:500) raised in rabbits against Us3 (kindly provided by Bernard Roizman) followed by anti-rabbit Alexa 594-conjugated secondary antibodies (1:500) (A11037; Thermo Fisher). After staining nuclei with 4',6-diamidino-2-phenylindol (DAPI; Roche, Mannheim, Germany), cells were embedded in glycergel mounting media (C0563; Dako North America, Carpinteria, CA, USA) and 25 mg/ml DABCO (1,4-diazabicyclo [2.2.2] octane; 33480, Fluka, Buchs, Switzerland) and analyzed using a confocal laser scanning microscope (SP2, Leica, Wetzlar, Germany).
Inoculated cells were harvested at 20 hpi and prepared according to (Tokuyasu, 1973; Tokuyasu, 1980). Cells fixed with 4% formaldehyde in 0.1 M Na-phosphate, pH 7.4, for 2 h at room temperature were scraped from the flasks, washed three times with 0.1 M Na-phosphate by centrifugation at 13,000g for 30 s, and pelleted in 12% gelatine by centrifugation at 13,000g for 3 min at 37°C. After gelation at 4°C, 1 mm3 blocks were immersed overnight in 2.3 M sucrose constantly rotating at 4°C. The infiltrated blocks were mounted on specimen holders, frozen by plunging into liquid nitrogen, and placed into the cryo-microtome (UC6, Leica, Vienna, Austria) at -120°C. Ultra-thin sections of 80–100 nm were collected on carbon-coated formvar films mounted on hexagonal 100 mesh/inch copper grids. Sections were washed by floating on several drops of buffer and blocking solutions prior to routine labeling procedure (Slot et al., 1991) with primary antibodies (1:30) against HA (SC-805, Santa Cruz Biotechnology), and secondary anti-rabbit antibodies (1:30) coupled to 12 nm colloidal gold (111-205-144, Jackson ImmunoResearch, West Grove, PA, USA). After labeling, sections were washed on distilled water droplets, stained by immersing in a mixture of 1.8% methyl cellulose and 0.4% uranyl acetate (Griffiths et al., 1983) for 10 min, and dried for imaging by TEM (Philips, CM12). For controls, primary antibodies were omitted, and labeling was also performed in wtHSV-1 infected cells.
Infection with HSV-1ΔUs3 induces formation of folds and invagination of nuclear membranes associated with accumulation of virions (Poon et al., 2006; Reynolds et al., 2002; Wisner et al., 2009). Infection with CK177ΔUs3gK-HA, a similar mutant lacking Us3 but equipped with an epitope-tagged gK, also induced multiple folds and invaginations of nuclear membranes containing virions. Moreover, homogenous, proteinaceous structures occurred within nuclei and cytoplasm of CK177ΔUs3gK-HA (Figure 1) and R7041ΔUs3 (Figure 2A) infected cells. Golgi complexes consisted of multiple stacks comprising thick electron dense membranes especially at the trans face (Figure 3) similarly as described in R7041ΔUs3 (Figure 2B) infections (Wild et al., 2015). We conclude that the absence of Us3 is responsible for the alterations in the Golgi architecture, while the epitope-tagged gK-HA has no obvious impact on Golgi disorganization. Unprocessed images of this, and all other figures, are available as Underlying data (Tobler et al., 2019).

The inner nuclear membrane (INM) (i) formed invaginations and multiple folds shown in detail in the inset. Capsids (c) bud at the INM, and virions accumulate in the perinuclear space delineated by the INM and outer nuclear membrane (ONM) (o), and to a much less extent in adjacent cisternae of the endoplasmic reticulum (RER). A few naked capsids (nc) are in close vicinity to Golgi membranes (Go). Note the presence of electron dense material (p) with the appearance of proteins in the nucleus and the cytoplasm. Bars, 200 nm.
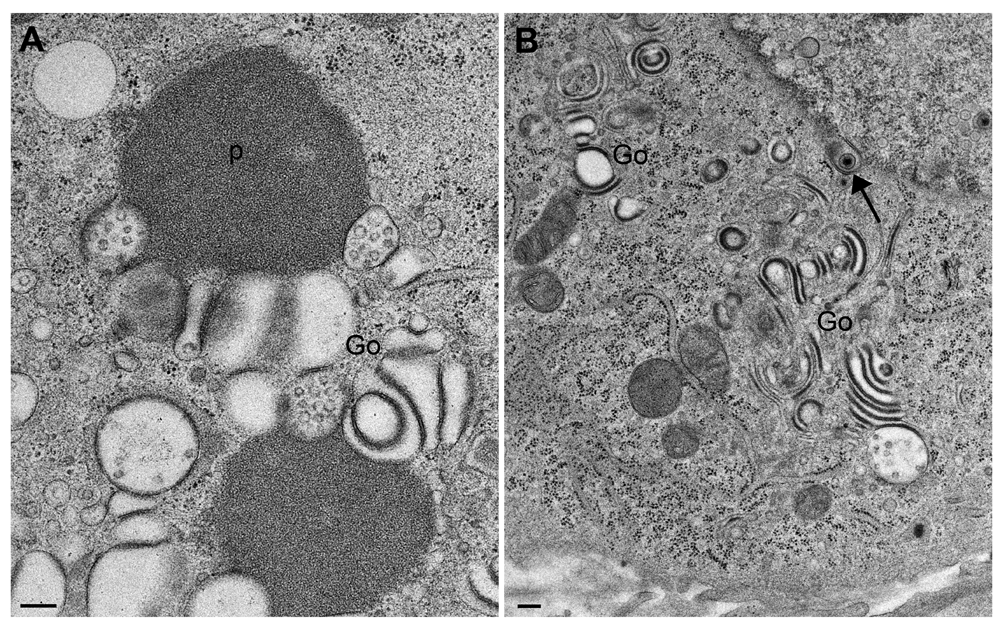
(A) homogenous, proteinaceous deposits (p) adjacent to altered Golgi membranes (Go), and (B) Golgi fields containing thick, electron dense membranes with the staining properties of proteins. Note the virion within double-coated structures (arrow), and the one in the endoplasmic reticulum (beside the arrowhead). Bars, 200 nm.
Transmission electron microscopy of Vero cells 24 hpi (A) and 20 hpi (B) with CK177ΔUs3gK-HA. The nucleus in (A) is tangentially sectioned so that invaginations of the inner nuclear membrane (arrow) appear as hibernations. The Golgi complex (Go) comprises multiple stacks consisting of electron dense membranes similar as in R7041ΔUs3 infections. One capsid (c) is in the cytoplasm, and two virions (one indicated by “v”) are within small concentric vacuoles. Note the well-preserved mitochondria (m). (B) shows details of Golgi membranes, which continue into rough endoplasmic reticulum membranes (thick arrow), and a tangentially sectioned Golgi field. Bars, 200 nm.
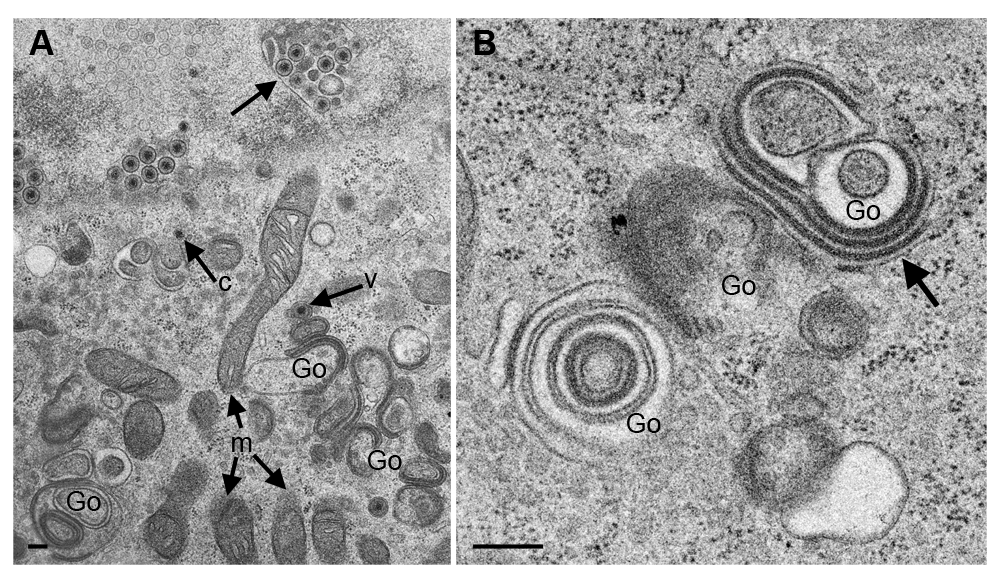
Budding of capsids at nuclear and Golgi membranes starts by insertion of budding proteins that appears in electron micrographs as a dense layer (Leuzinger et al., 2005). The major budding proteins at nuclear membranes are UL31/UL34 (Bigalke et al., 2014; Hagen et al., 2015), while gK and UL20 protein are responsible for budding at Golgi membranes (Melancon et al., 2007). Infection with CK177gK-HA revealed that gK localizes in cytoplasm, nuclear rim and even in nuclei (Figure 4A) by immunofluorescence using antibodies against HA. Following infection with CK177ΔUs3gK-HA, gK-HA signals are strongly enhanced in the cytoplasm (forming large clusters), at the nuclear rim and in nuclei (Figure 4B). At the ultrastructural level, Golgi membranes (Figure 5), nuclear membranes and viral envelopes (Figure 6) and the homogeneous structures in nuclei and cytoplasm (Figure 7) were distinctly labeled in CK177ΔUs3gK-HA infected cells. Immunogold labeling agrees with the distribution of gK-HA visualized by immunofluorescence implying that gK was, in addition to Golgi localization, translocated into nuclei, incorporated into nuclear membranes, and became part of the viral envelope during budding.

Immunofluorescence microscopy of Vero cell at 20 hpi with CK177gK-HA (A) or CK177ΔUs3gK-HA (B). CK177gK-HA infected cells show fine but dense distribution of gK-HA over the entire cytoplasm, at the nuclear rim and, at a less extent, in the nucleus. Distribution of Us3 is similar though more intense at the nuclear rim. In the absence of Us3, gK-HA signals are markedly enhanced in all three compartments. gK-HA partly localizes with wheat germ agglutinin (WGA) as a marker for Golgi membranes. The fate of the Golgi complex is under investigation using specific markers.

Gold particles are irregularly distributed on electron dense Golgi membranes (Go) as well as on proteinaceous deposits (p). Bars, 200 nm.
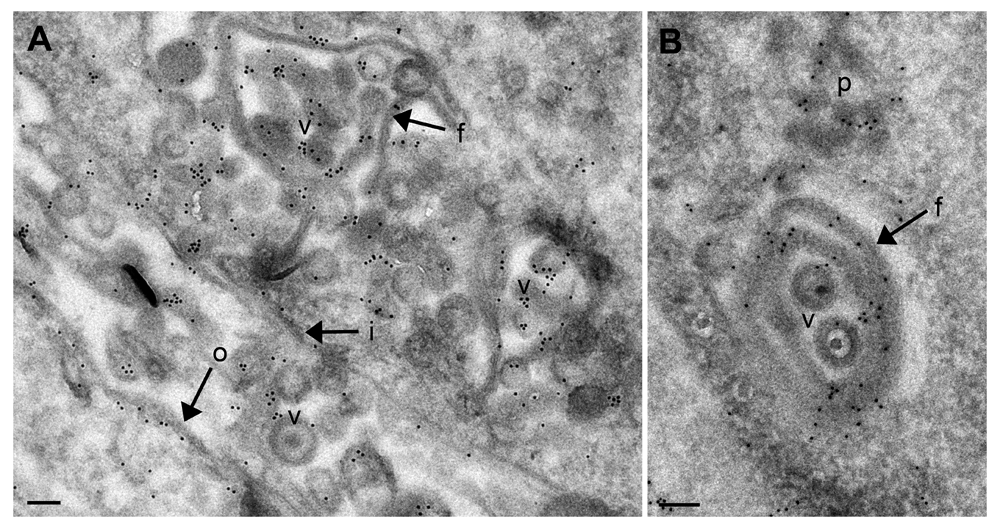
(A) Shows an overview. (B) Shows a detail of membrane folds protruding into the nucleus. Gold particles are located on nuclear membrane folds (f), virions (v), nuclear membranes (i, o) and on protein deposits (p) in the nucleus. Bars, 200 nm.
The precise functions of Us3 kinase has been reviewed by Kato & Kawaguchi (2018). However, the mechanism of regulation phospholipid-biosynthesis is unknown (Wild et al., 2012). Membrane enlargement in the absence of Us3 is associated with structural alterations including folding of nuclear membranes and malformation of Golgi stacks, respectively, and insertion of proteins into nuclear and Golgi membranes. Immunolabeling using antibodies against the HA tag recognized gK to be one component of altered membranes suggesting that deposition/insertion of gK is related to membrane overproduction in the absence of Us3.
HSV-1ΔUs3 virions are infective even though they are not released from the PNS, and do not pass the Golgi complex. gK has been reported to be involved in viral envelopment by Golgi membranes, not by nuclear membranes (Melancon et al., 2005). gK is essential for infectivity playing a significant role in viral entry (Foster et al., 2001; Musarrat et al., 2018). Therefore, gK must be provided during budding of HSV-1ΔUs3 at nuclear membranes. The intense labeling of gK on nuclear membranes and viral envelopes clearly demonstrates that gK becomes part of the viral envelope during budding of HSV-1ΔUs3. gK was also found in nuclei in both HSV-1ΔUs3 and CK177gK-HA infected cells indicating that gK is transported into the nucleus in the presence or absence of Us3. Indeed, others have also observed gK incorporated into nuclear membranes in the context of wtHSV-1 infection (Rajcani & Kudelova, 1998) suggesting that gK may also play a significant role in envelopment of wtHSV-1 at nuclear membranes.
Without Us3, Golgi and nuclear membranes proliferate in association with incorporation of gK that also localizes on virions remaining stuck in the PNS but mature to infectivity suggesting that i) Us3 kinase is involved in regulation in nuclear and Golgi membranes assembly possibly in association with gK synthesis and/or distribution, ii) gK may be the target of Us3 phosphorylation, which is needed for virions to proceed out of the PNS, and iii) gK may also be involved in nucleus-to-cytoplasm translocation in wtHS-1 because gK localizes on nuclei and nuclear rim in the presence of Us3. We hypothesize that Us3 interplays with mechanisms regulating synthesis and arrangement of Golgi membranes, nuclear membranes and gK, which might be the most significant role of Us3 remaining to be investigated.
Figshare: The herpes simplex virus 1 Us3 kinase is involved in assembly of membranes needed for viral envelopment and in distribution of glycoprotein K. https://doi.org/10.6084/m9.figshare.8131241.v2 (Tobler et al., 2019).
This project contains the raw images captured for each Figure, with the number of the Figure indicated on each file name.
Data are available under the terms of the Creative Commons Attribution 4.0 International license (CC-BY 4.0).
This study was supported by the Foundation for Scientific Research at the University of Zürich, Switzerland.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We thank Bernard Roizman, University of Chicago, for R7041ΔUs3 and antibodies against Us3.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Herpesvirus replication and pathogenesis
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
No
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
No
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: molecular virology and viral pathogenesis, herpesviruses and poxviruses.
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
References
1. Foster TP, Rybachuk GV, Alvarez X, Borkhsenious O, et al.: Overexpression of gK in gK-transformed cells collapses the Golgi apparatus into the endoplasmic reticulum inhibiting virion egress, glycoprotein transport, and virus-induced cell fusion.Virology. 2003; 317 (2): 237-52 PubMed AbstractCompeting Interests: No competing interests were disclosed.
Reviewer Expertise: Molecular virology and immunopathogenesis of herpes viruses
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |||
|---|---|---|---|
| 1 | 2 | 3 | |
|
Version 1 23 May 19 |
read | read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)