Keywords
herpes simplex virus 1, renin angiotensin aldosterone system, angiotensin converting enzyme-1, captopril, antiviral
herpes simplex virus 1, renin angiotensin aldosterone system, angiotensin converting enzyme-1, captopril, antiviral
Herpes simplex virus 1 (HSV-1) is a neurotropic virus that infects over 60% of the United States population and 90% globally1. HSV-1 is responsible for a diverse array of biological effects and physical manifestations, including cold sores and encephalitis. HSV-1 remains the leading infectious cause of corneal blindness and encephalitis2,3. Moreover, HSV-1 infection is a significant contributing factor in Human Immunodeficiency Virus (HIV) transmission and pathology3. Although HSV-1 infection is common, there is neither vaccine nor cure. Over 75% of HSV-1 antiviral drugs are nucleoside analogues that inhibit virus genome replication. Antivirals are effective in reducing the duration and frequency of outbreaks, but their efficacy is limited to active infections and doesn’t mitigate reactivation, which occurs from latent infections. Current therapeutic targets have major limitations, such as the requirement for an active infection, high dosing frequencies, and the development of antiviral resistance. Targeting a common mechanism of action contributes significantly to the development of antiviral resistance, an emerging problem for treating HSV-1 and other virus infections. The poor bioavailability of most HSV-1 antivirals requires high dosing frequencies, up to three to five times per day, to maintain therapeutic levels. However, Valacyclovir, a pro-drug of acyclovir, is the exception with an improved bioavailability profile compared to its parent drug and thus allows a reduced dosing frequency. Acyclovir, the mainstay of treatment for HSV-1 infection, consists of a synthetic nucleoside analogue that targets viral DNA polymerase and functions by inhibiting viral replication due to its affinity for viral thymidine kinase4,5. Acyclovir’s clinical use in immunocompromised patients can lead to the development of antiviral resistance6. HSV-1-induced pathologies, the global burden that results from HSV-1 infection, and the lack of effective treatments highlights a need for the identification of novel antiviral strategies and therapeutics. Thus, there is an urgent medical need to identify novel drug targets and subsequently develop and evaluate novel therapeutic strategies and interventions that can exert their action through a distinct mechanism.
Components of the Renin-Angiotensin Aldosterone System (RAAS), known for their function in the homeostatic control of arterial blood and osmotic pressures, have been implicated in regulating virus activity. Human cytomegalovirus, Human Herpesvirus-5 (HHV-5), has been demonstrated to cause an increase in arterial blood pressure7, further supporting our investigation into the relationship between viral infections and the RAAS system. The ongoing Severe Acute Respiratory Coronavirus 2 (SARSCoV2)-induced pandemic further highlights the interaction between viruses and RAAS components as it utilizes Angiotensin Converting Enzyme-2 (ACE-2), a cellular protein, as its primary entry receptor8. These findings support our hypothesis that components of RAAS may attenuate HSV-1 infection. Losartan, an angiotensin type 1 receptor blocker, and des-aspartate-angiotensin 1, both inhibitors of RAAS, have demonstrated antiviral effects in coxsackievirus and rhinovirus infections, respectively9,10. The work described here evaluated the hypothesis that Angiotensin Converting Enzyme-1 (ACE-1) inhibition is protective against HSV-1-induced cytopathic effects (CPE). We have demonstrated that the ACE-1 inhibitor captopril attenuates CPE in HSV-1-infected SH-SY5Y neuroblastoma cells. The results reported here are the first, to our knowledge, to show that captopril protects neuroblastoma cells from HSV-1-induced CPE by impairing virus genome replication.
The neuroblastoma SH-SY5Y cell line (ATCC, CRL2266, lot # 11C016) was purchased from Sigma-Aldrich (St. Louis, MO). Cells were maintained in Dulbecco's Modified Eagle's Medium (DMEM; Sigma Aldrich, St. Louis, MO) supplemented with 10% fetal bovine serum (Sigma Aldrich, St. Louis, MO) and 7% Penicillin and Streptomycin (Sigma Aldrich, St. Louis, MO). Cells were seeded into flasks containing supplemented medium and maintained at 37°C in saturated humidity with 5% CO2. For assays, SY5Y cells were sub-cultured into 96-well plates at a seeding density of 1× 105 cells per well.
The F strain of HSV-1 was graciously contributed by the Fraser Lab at The University of Pennsylvania. Cells were infected at a multiplicity of infection (MOI) of 0.01. During infection, virus was delivered to cells in serum-free DMEM and cells were administered either captopril (Sigma Aldrich, St. Louis, MO; administered at various concentrations (0, 1pM, 10pM, 1nM, 10nM, 1µM)) or vehicle control (0.1% dimethyl sulfoxide (DMSO)) post-infection. One-hour post-infection, inoculum was aspirated and an equal volume of complete media was added to the cells. Cells were incubated until the cell viability assay or imaging analysis was performed. Andrea Bertke (Virginia Tech) donated the green fluorescent protein-tagged HSV-1 (GFP-HSV-1).
Cell viability was quantified via the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium (MTT) assay (Sigma-Aldrich, M5655). Cells were cultured with cDMEM in 96-well plates until they were 90% confluent. Next, subsets of wells were infected with HSV-1 (F strain) at a MOI of 0.01 for 24 hours. MTT solution (5mg/ml) was added to each well and incubated for four hours at 37°C. After removing the supernatant from wells, 100 µl DMSO was added to each well. The absorbance was read at 570 nm with a 630nm reference wavelength via a Benchmark Plus (BIO RAD) spectrophotometer. All experiments were repeated in triplicate and processed in parallel.
Time course analysis was evaluated via the previously described methods used in the cell viability protocol, utilizing the MTT assay at 3, 9, and 24 hpi. All experiments were repeated in triplicate and processed in parallel.
Photomicrographs were obtained utilizing an Olympus IX81 microscope equipped with a Hamamatsu camera. MetaMorph software (version 6) was used to image cells, and photomicrographs were taken with the differential interference contrast (DIC) filter at a final magnification of 20x (Micro-Manager is a free alternative if access to MetaMorph is not available). Three distinct fields of view were imaged per well.
Green fluorescent protein-tagged HSV-1 (GFP-HSV-1) was used to infect cells, as previously described. There were three treatment groups: Mock, Infected/Vehicle, and Infected/Captopril. A Mock/Captopril group was not used in this experiment as we wanted to examine the effect of captopril of HSV-1 replication and previous data demonstrated that captopril was non-toxic. Photomicrographs were taken with a DIC filter at 8, 24, and 48 hours post infection (hpi).
AutoDock Vina (version 1.1.2), a docking program, was used to investigate the interaction between HSV-1 glycoprotein D (gD) (Protein Data Bank (PDB) ID: 1P7C) and captopril (PDBID: 2X8Z). Captopril was collected from the bound crystal structure of AnCE using PyMOL V1.8.6.2. AutoDock Vina was used to approximate the favorability of HSV-1 gD binding with captopril. The crystal structure of both molecules were obtained from the PDB and used to generate 3D structures. To perform the docking simulations, AutoDock Vina generated a grid box to enclose the active site with dimensions of 126 Å × 126 Å × 126 Å and a grid spacing of 0.375 Å using Autodock Tools. The results are represented by free energy binding.
To render the Ligand Interactions Diagram, Schrodinger Maestro V10.6 was used. Initially, crystal structure of HSV1 gD (receptor) and captopril (ligand) were obtained from the PDB as mentioned above. The imported protein structure obtained from the PDB was not suitable for immediate use in the molecular docking study as it contained excess material (i.e., water molecules, metal ions, cofactors, etc.). To overcome this obstacle, the protein structure was prepared using the protein preparation wizard (preprocessed, optimized, and minimized) in Maestro v10.611,12. The ligand structures of the data set were prepared by LigPrep module of Schrodinger v10.612.
SH-SY5Y neuroblastoma cells were treated with either vehicle (0.1% DMSO), the F strain of HSV-1 (MOI=0.01), or captopril and HSV-1 (F strain, MOI=0.01) together in media without serum for one hour. After this, an equal volume of complete media was added to the cells. Next, RNA was isolated from treated cells according to the manufacturer’s instructions (Qiagen AllPrep DNA/RNA Mini Kit; Cat. No. 80204, Lot # 154018161). Following RNA isolation, the purity and concentration of resulting RNA were measured using the Thermo Scientific Nano Drop 2000 Spectrophotometer. All samples were stored at -80°C for future use.
cDNA synthesis. The protocol used to synthesize first-strand complementary DNA from purified RNA was followed using the Invitrogen SuperScript III First-Strand Synthesis System for RT-PCR (Thermo Fisher, Cat. No. 18080-051). RNA (100ng) isolated from SH-SY5Y cells was placed in Bio-Rad Low-Profile PCR Tubes containing 10mM dNTP mix and 50ng of random hexamer primers. The final volume in each tube was 10 uL. Positive (HeLa RNA) and negative control (tube containing HeLa RNA but no reverse transcriptase) reactions were also prepared. Samples were placed in a Bio-Rad T100 Thermal Cycler and incubated at 65°C for 5 minutes, cDNA synthesis mix (according to the manufacturer’s instructions) was added to each tube including the positive and negative controls. After following subsequent steps detailed in the SuperScript III manual, the resulting cDNA was stored at -20°C.
qPCR cycle settings. The protocol for qPCR was adapted from Thermo Fisher Scientific PowerUp SYBR Green Master Mix (Catalog No., lot# 1601009). Reactions were run in triplicate in Bio-Rad 96-well PCR plates. Primers for each gene of interest were synthesized by Integrated Data Technologies and diluted to 10uM concentrations with DEPC-treated water. Human GAPDH was used as a reference gene, and ICP0 was utilized to measure HSV-1 viral gene expression. The primer sequences for human GAPDH were: 5’ TGG GCT ACA CTG AGC ACC AG 3’ (forward) and 5’ GGG TGT CGC TGT TGA AGT CA 3’ (reverse). The forward primer sequence for ICP0 was 5’ CCC ACT ATC AGG TAC ACC AGC TT 3’. The sequence for ICP0 reverse primer was 5’ CTG CGC TGC GAC ACC TT 3’. Each analyzed well of the PCR reaction plate contained the following: forward primer (500nM), reverse primer (500nM), PowerUp SYBR Green Master Mix (2X), DEPC-treated water, and cDNA (10ng). The PCR plate was sealed and centrifuged, and inserted into the CFX96 Real-Time System. The qPCR cycling mode was as follows: two minutes at 50°C followed by two minutes at 95°C. Next, a total of 40 denature (15 seconds at 95°C) and anneal (60°C) cycles were utilized. Melt curve analyses were performed with each reaction. ICP0 gene expression levels were normalized to human GAPDH levels.
All analyses for statistically significant differences were performed utilizing a two-way analysis of variance (time x infection status or concentration x infection status). The alpha value was set at 0.05. If warranted, a Tukey’s post-hoc test was performed. For the concentration-response curve, a Dunnett’s multiple comparisons post-hoc test was used, comparing the different concentrations to the vehicle (0) control. Data are expressed as means (± SEM). GraphPad Prism (version 6) statistical analysis software was utilized to perform statistical comparisons and prepare graphical representations of quantitative data.
To determine the concentration dependent response of captopril on SH-SY5Y neuroblastoma cells, we first assessed cell viability and cellular morphology via the MTT assay and DIC microscopy, respectively. SH-SY5Y cells were treated with a range of concentrations (0, 1pM, 10pM, 1nM, 10nM, 1uM) of captopril. To investigate if captopril treatment alone adversely affected cell viability, the experiment was first performed in uninfected cells. The concentration response data in uninfected SH-SY5Y cells demonstrated that captopril (Figure 1) did not impact cell viability as viability remained approximately 100% with no statistically significant differences observed at any concentration.
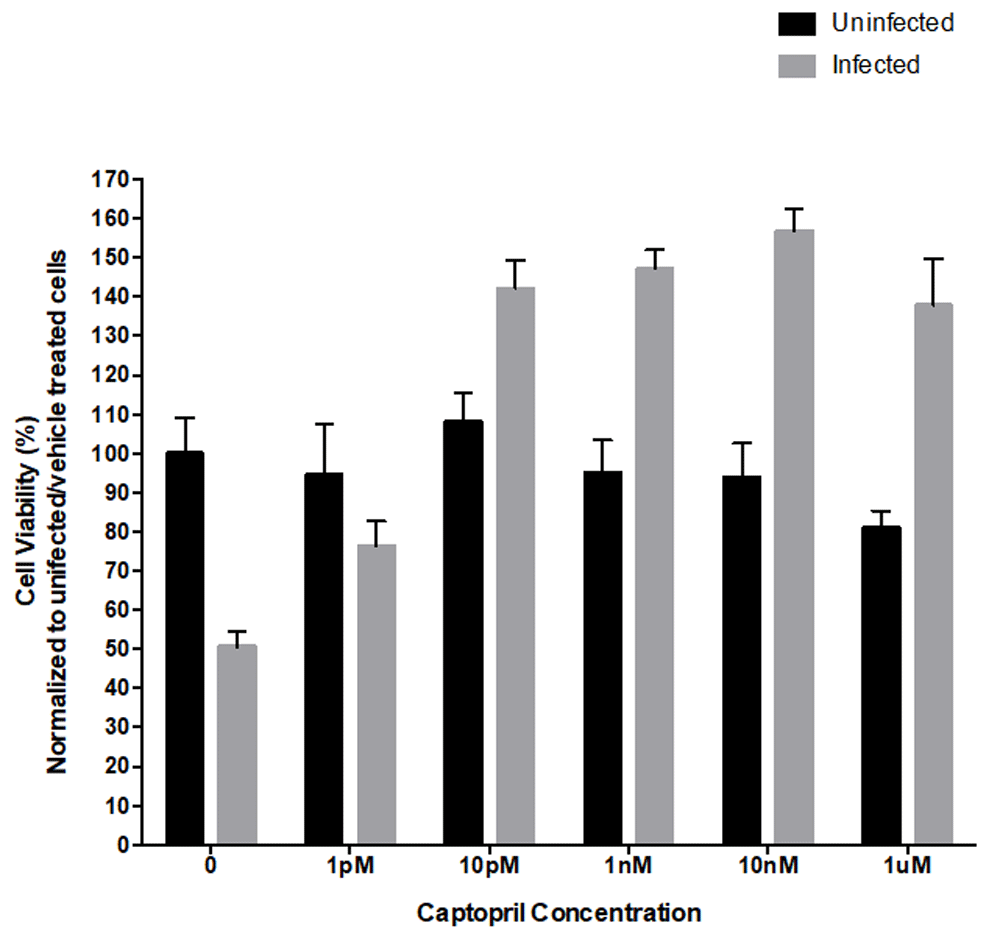
The response of uninfected and HSV-1 infected SH-SY5Y neuroblastoma cells to varying concentrations (0, 1pM, 10pM, 1nM, 10nM, 1uM) of captopril was measured via MTT cell viability assay at 24 hours post infection and treatment. Data from uninfected and captopril treated cells showed no significant change in cell viability. Cells infected with the F strain of HSV-1 (MOI=0.01) demonstrated a statistically significant reduction in viability, compared to their uninfected counterparts (F(1,98)=10.22, p < 0.005). HSV-1 infected, captopril-treated cells had a statistically significant, dose-dependent increase in cell viability from 10pM to a maximum efficacy observed at 10nM, compared to infected, untreated cells (F(6,98)=23.82, p<0.0001). The asterisk denotes that the HSV-1-infected group was statistically significant from the uninfected group at the 0 concentration. The plus signs represent groups (infected with HSV-1 and treated with captopril) statistically significant from their counterparts that were infected the F strain of HSV-1 but received no captopril treatment. Each bar represents the average of six cell culture replicates.
Alongside the uninfected cells, HSV-1 infected SH-SY5Y cells were treated with a range of concentrations of captopril (0, 1pM, 10pM, 1nM, 10nM, 1uM). Results from the MTT cell viability assay demonstrated captopril treatment caused a significant increase in viable cells at 10pM, 1nM, 10nM, and 1μM (F(6,98)=23.82, p<0.0001), when compared to the infected, untreated group. In subsequent experiments, we chose to use the 10nM dose of captopril as our results indicated it offered maximum protection, compared to the other doses evaluated, and was less than the 1μM dose.
Experiments were conducted to measure cell viability at different time points (3, 9, and 24 hpi). Cell viability measurements were equivalent among all the groups at 3 hpi. At 9 hpi (F strain of HSV-1, MOI=0.01), the Virus/Vehicle group had 20% fewer viable cells (F(3,8) =23.27; p<0.0005) compared to every other treatment group (Figure 2). At 24 hpi there were approximately 25% less viable cells in the Virus/Vehicle group compared to all other groups. At 9 and 24 hpi, the only significant difference observed was between the Virus/Captopril group and the Virus/Vehicle group. Differences due to time alone were not statistically significant.
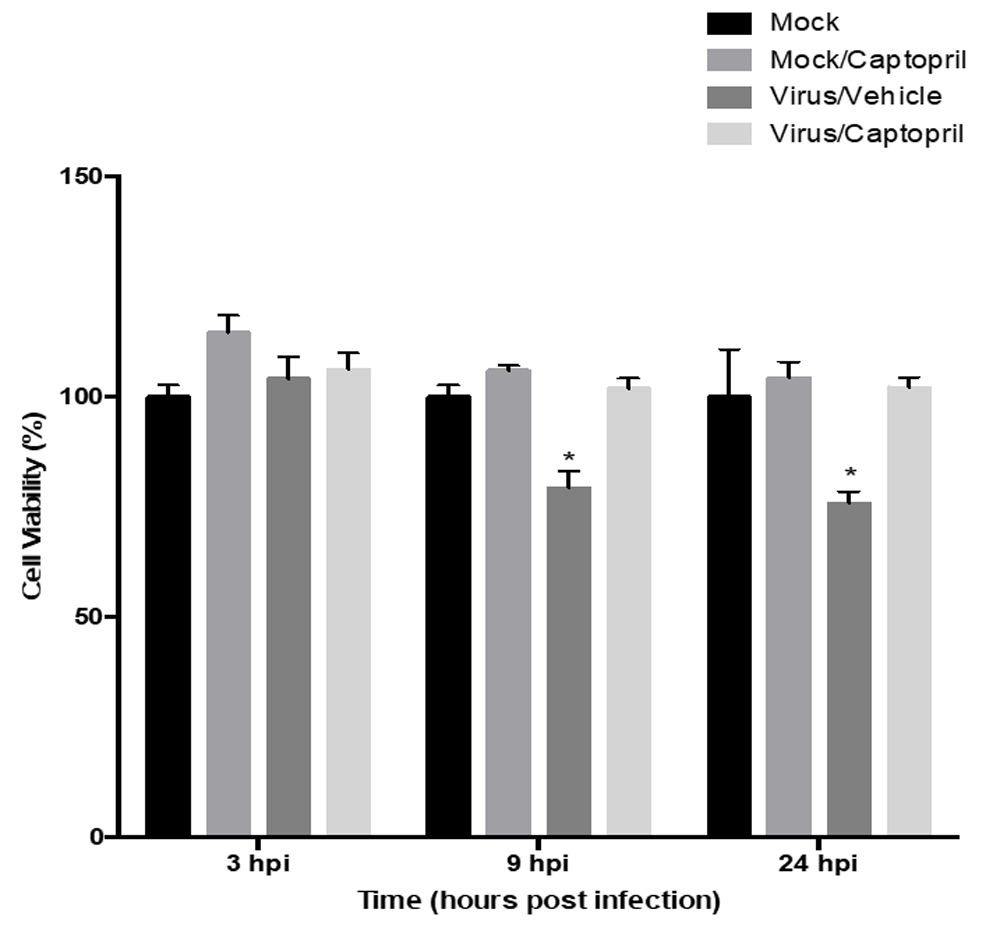
Cell viability was evaluated via MTT assay at 3, 9, and 24 hours post infection (hpi) with the F strain of HSV-1 (MOI=0.01). At 9 and 24 hpi, the Virus/Vehicle group was statistically significantly different compared to the all other groups. Each bar represents the average of six biological replicates. The asterisks indicate groups that were statistically significant from the other groups (not denoted with an asterisk) at their respective time points.
Photomicrographs were taken as a qualitative analysis to corroborate the MTT results (Figure 3). The experimental groups (Mock, Mock/Captopril, Virus/Vehicle, and Virus/Captopril) were exposed to the same conditions (24 hpi and 10 nM captopril) prior to obtaining photomicrographs. Cells in Figure 3a (Mock) are representative of healthy SH-SY5Y cells indicated by the confluence and the appearance of intact cellular extensions. Cells in the Mock/Captopril group (Figure 3b) were also very confluent and exhibited similar structural characteristics to the Mock group (Figure 3a). In both of these groups, there were no indicators of substantial cell death such as changes in cellular morphology (i.e., cell rounding) and loss of anchorage dependence (i.e., floating cells). HSV-1 infected cells (Virus/Vehicle) exhibited a reduction in confluence (Figure 3c). Cells in this group also demonstrated multiple instances of cytopathy such as changes in morphology, the loss of anchorage dependence, and a decrease in neurite extensions on cells that remained attached. HSV-1 infected cells treated with captopril (Virus/Captopril, Figure 3d) were more confluent compared to the Virus/Vehicle group and maintained normal characteristic extensions of neuroblastoma cells.

Photomicrographs show morphological changes 24 hours after captopril (10nM) administration. Uninfected cells administered vehicle (0.1% DMSO) (3a) or captopril alone (3b) were indistinguishable in confluence and structure. In the absence of captopril, HSV-1 infected cells were rounded, an indicator of adverse cell health (3c). Captopril treated, HSV-1 infected cells (3d) maintained their neurite extensions (as seen in 3a and 3b) and were more confluent when compared to HSV-1 infected, untreated cells (3c). (Arrows are intended to illustrate morphological changes.) Additionally, cells treated with captopril preserved the neurite extensions associated with healthy SH-SY5Y neuroblastoma cells. Images were taken at a final magnification of 200x using an IX Olympus 81 inverted microscope and MetaMorph microscopy imaging software.
Cells infected with a GFP-expressing HSV-1 strain were analyzed at 1, 3, 8, 24, and 48 hpi. In the Virus/Vehicle group, GFP expression was not observed until 8 hpi. After this time point, the percentage of GFP-positive cells increased in a time-dependent manner. At 8 hpi, an average of 13% of cells were GFP-positive in the Virus/Vehicle group compared to only 1% in the Virus/Captopril group (F (7,11) =3.58; p<0.05; Figure 4.). At 24 hpi, 30% of cells were GFP-positive in the Virus/Vehicle group compared to 25% of GFP-positive cells in the Virus/Captopril group. This pattern continued at 48 hpi, with 61% of GFP-positive cells observed in the Virus/Vehicle group versus 27% of GFP-positive cells in the Virus/Captopril group (F (7,11) =2.72; p<0.05).
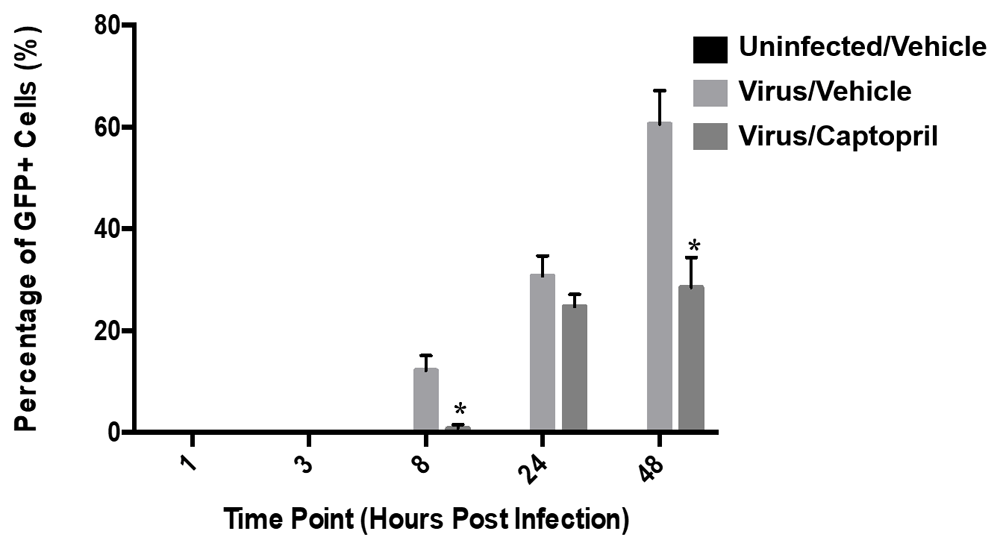
HSV-1 used was constructed to express GFP as a measure of replication. The percentage of GFP positive cells was quantified via manual image analysis at 1, 3, 8, 24, and 48 hpi. At 1 and 3 hpi there were no GFP-positive cells in any of the treatment groups. At later time points (8, 24, and 48 hpi), there was a time-dependent increase in percentage of cells expressing GFP in both the Virus/Vehicle and Virus/Captopril group. Each bar represents the average of six biological replicates. The statistically significant differences in the percentage of GFP positive cells were observed between the Virus/Vehicle and the Virus/Captopril groups at 8 and 48hpi (F(2,30) =12.82; p<0.0001).
Micrographs of cells infected with the GFP-HSV were imaged at 20X magnification. Cells receiving the Uninfected/Vehicle treatment did not exhibit any GFP fluorescence at any time point (Figure 5a–c). With the administration of the GFP-expressing virus (MOI=0.01), cytopathic effects (e.g., cellular rounding) were noted as well as fluorescent imaging at all three time points (8, 24, and 48 hpi; Figure 5d–f). Co-administration of captopril (10nM) with the GFP-HSV reduced the occurrence of both cytopathy and green fluorescence (Figure 5g–i).
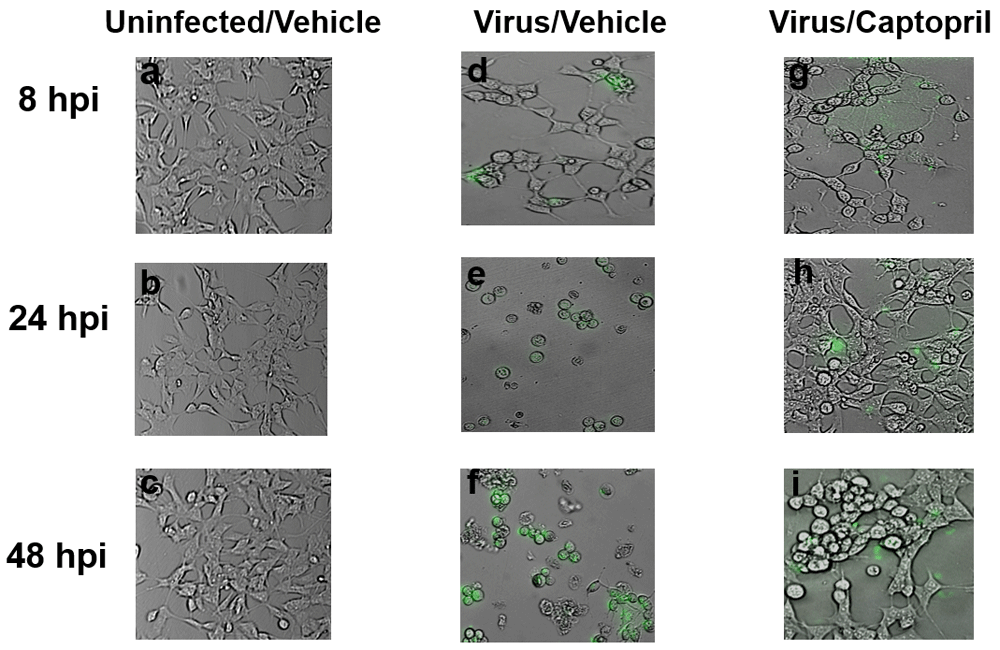
Photomicrographs from cells in the Uninfected/Vehicle, Virus/Vehicle, and Virus/Captopril groups were imaged at a final magnification of 200X using an Olympus IX81 microscope. An HSV-1 virus fused with GFP was utilized in this experiment. GFP expression, an indicator of viral replication, was not detected in any cells in the Uninfected/Vehicle group (a–c). GFP expression increased from over time in cells infected with GFP-HSV (MOI=0.01, d–f). With co-administration of captopril (10nM), there was a decrease in green fluorescence and dying cells (indicated by the rounded morphology) in HSV-1 infected cells (g–i).
We conducted molecular modeling studies to determine the free binding energy (∆G) between the ACE inhibitor captopril and HSV-1 gD. Results demonstrated that the free binding energy between captopril and HSV-1 gD was favorable and calculated to be -3.9 kCal/mol (Figure 6A). Further evaluation of the interaction between HSV-1 gD and captopril identified the specific amino acid residues responsible for the interaction (Figure 6B).

(A) A ribbon diagram of HSV-1 gD is shown in complex with captopril (small molecule). Molecular modeling via AutoDock Vina determined that captopril has a favorable interaction with HSV-1 gD, indicated by a predicted free binding energy (ΔG) of -3.9 kCal/mol. (B) Captopril-HSV-1 gD predicted interaction with specific amino acid residues of gD color coordinated by charge.
Transcripts of the viral gene ICP0 were detected at 2 hpi by SH-SY5Y neuroblastoma cells infected with the F strain of HSV-1 (Virus/Vehicle Group). qPCR results demonstrated that captopril co-administration to HSV-1 infected cells decreased ICP0 gene expression by approximately 34%, compared to infected cells not administered captopril (p<0.0001; Figure 7).
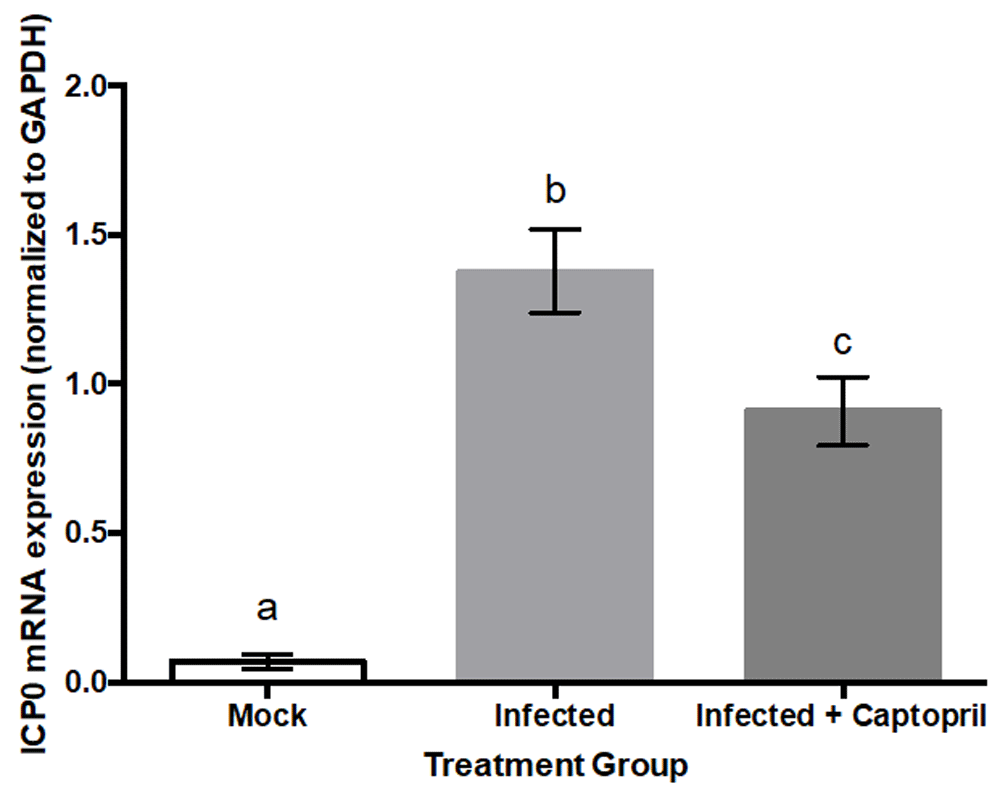
Two hours after being infected with the F strain of HSV-1 (MOI=0.01), cells in the Virus/Vehicle group had transcript levels of the viral gene ICP0 statistically significantly greater than those in the Uninfected/Vehicle condition (F(2,23)=41.59; p<0.0001). ICP0 was not detected in uninfected SY5Y neuroblastoma cells. Each treatment group consisted of eight biological replicates. qPCR was executed in a parallel fashion and each bar represents the average of three replicates. Transcript levels of the viral gene ICP0 were normalized to levels of human GAPDH.
To strengthen the work relating RAAS components and antiviral activity, we studied the anti-HSV-1 effects of captopril in SH-SY5Y neuroblastoma cells. Based on previous findings from Zhang et al.9, we hypothesized that ACE inhibition would confer protective effects induced by HSV-1. We have demonstrated that captopril attenuates HSV-1induced CPE in SH-SY5Y cells. Results of cell viability assays indicate captopril protects cells from HSV-1-induced cellular death by 18% when compared to untreated counterparts. This finding was further substantiated by a qualitative assessment of photomicrographs in which GFP fluorescent intensity was used as a marker for virus entry and replication. Experiments using GFP-HSV-1 and subsequent qt-PCR demonstrated that captopril blunted HSV-1 replication. These results support our hypothesis that captopril reduces the cytopathic effects and replication of HSV-1.
Infection with HSV-1 results in life-long infection with initial infection typically occurring in childhood. HSV-1 can cross the blood brain barrier, promote inflammation and damage the central nervous system. There are few drugs licensed for the treatment of HSV-1 infections. Acyclovir remains the “gold standard” in HSV-1 therapy, more than thirty years after its development4. Extensive clinical use of acyclovir and other approved treatments has contributed to the emergence and persistence of resistant viral strains, particularly in immunocompromised individuals13. Treatments effective against HSV-1 are nucleoside analogues with the exception of foscarnet, a pyrophosphate analogue. Foscarnet is primarily restricted to individuals whose virus infections are resistant to acyclovir14. Thus, few treatment options coupled with the emergence of resistant viral strains contribute to a growing need of novel antiviral targets and strategies13–15. Components of the RAAS have been implicated and shown to play a role in regulating virus activity and select RAAS inhibitors cross the blood brain barrier, leading us to hypothesize that captopril could attenuate the cytopathic effects of HSV-1 in SH-SY5Y cells9,10.
SARS-CoV-2 that caused a worldwide pandemic also has links to RAAS components. SARS-CoV-2 utilizes ACE-2 as a receptor to facilitate entry into host’s cells8. This finding further demonstrates the potential significant relationship between RAAS components and viruses.
In this study, we investigated the role of ACE-1 and its inhibitors in HSV-1 replication. We determined the most effective captopril concentration for reducing the cytopathic effects of HSV-1 in SH-SY5Y cells was 10nM. According to Henda et al.16, captopril has an IC50 range of 1.79-15.1nM when used with synthetic substrates and an IC50 around 16.71µM when used with Angiotensin I. Thus, the concentration needed to reduce HSV-1 replication is well below its active range in inhibiting ACE. Other RAAS inhibitors, such as losartan, have been evaluated in vivo for antiviral effects using a concentration of 12mg/kg17,18 against coxsackievirus B3 which induces myocarditis. Des-aspartate-angiotensin I, an angiotensin nonapeptide, has also been shown to have an antiviral effect against rhinovirus when used between a 10-10 and 10-12 nanomolar range10. At such a low concentration, 10nM, we do not anticipate any significant non-specific effects. Collectively, our quantitative and qualitative data indicates that captopril does not significantly alter cell viability or induce any visual cytopathic effects in uninfected cells.
Time course experiments were conducted at 3, 9, and 24 hpi to evaluate if there were time-dependent changes in cell viability. Time points were selected based on reported times for the initiation of viral gene expression. Immediate-early (α) genes, such as infected cell protein 0 (ICP0), are typically expressed within 1 hpi; early genes (β), such as thymidine kinase, are usually expressed within 3hpi; and late (γ) genes, such as glycoprotein C (gC), are typically expressed between six and eight hpi; and Latency Associated Transcripts are expressed upon initial infection and are continuously abundantly expressed following initial infection. Based on relevant literature and our preliminary findings, we decided to observe the effect of viral infection at 24 hpi. Significant differences were observed between the Virus/Vehicle group when compared to the Virus/Captopril groups at 9 and 24 hpi.
Computational approaches to drug discovery are used extensively to determine the potential of candidate and novel drugs. The current COVID-19 crisis reinforces the use of computational approaches in drug discovery. We used a computational approach to evaluate the interaction between ACE inhibitors and HSV-1 via molecular modeling and found that captopril favorably interacts with HSV-1 gD with a negative free binding energy (ΔG) of -3.9 kCal/mol while the other evaluated ACE inhibitors (lisinopril and fosinopril) do not interact with HSV-1 gD.
The primary limitation to this study is the use of only one cell line. Future studies to determine the effects of captopril in alternative cell lines infected with HSV-1 are warranted. Moreover, it would be prudent to investigate how other methods of ACE inhibition or Angiotensin II receptor antagonism affect HSV-1 replication. Future work will also determine how well captopril impairs HSV-1 replication at higher MOIs. Yet, the work detailed here provides a firm basis for captopril to be further evaluated as a novel antiviral effective against HSV-1.
The overall goal of this study was to evaluate the anti-HSV-1 properties of captopril. Our results, both quantitative and qualitative in nature, demonstrate that captopril exerts a protective effect against HSV-1. Thus, these results strengthen work addressing antiviral effects of RAAS components and offers potential for a novel strategy for combatting HSV-1 infection. As a commonly prescribed and relatively safe blood pressure medication, captopril has demonstrated promising potential as a therapeutic intervention for the treatment of HSV-1. Problems associated with current HSV-1 treatment regimens are largely centered around antiviral resistance and high dosing frequencies due to low oral bioavailability. Acyclovir is reported to have a bioavailability between of 20%, requiring a dosing frequency of five times per day19 to maintain therapeutic levels. Valaciclovir, a prodrug of acyclovir, also used for the treatment of herpes virus infections increases bioavailability to 55%20, which causes a decrease in dosing frequency from five to three times per day to be effective, but is accompanied by a price increase which may affect which treatment options are prescribed. Captopril has an oral bioavailability of approximately 65%21, which if effective as an antiviral, as data suggest, may result in a further decrease in dosing frequency, potentially leading to an increase in patient compliance and is relatively inexpensive. Additionally, captopril crosses the blood brain barrier (BBB), and thus could decrease the CNS inflammatory response and prevent further damage caused by viral-mediated BBB permeability.
Collectively, our findings warrant further studies that will test the mechanisms underlying the antiviral effect demonstrated in this work. Future work will delineate any possible direct interactions between the drug and HSV-1 itself as well as any effects of captopril on viral entry. This work highlights a critical area of study that could provide a novel approach to reducing HSV-1 entry and reducing the pathologies associated with its infection and reactivation while providing a much-needed novel therapeutic alternative.
Current treatment options for HSV-1 and HSV-1-associated neurological complications largely rely on high dosing frequencies of nucleoside analogues, such as acyclovir, famciclovir and ganciclovir, which can have damaging side effects on multiple organ systems20,21. Previous studies have made connections between RAAS and viral activity, demonstrating that RAAS components may have antiviral abilities13,16. Here, we studied antiviral properties of the ACE inhibitor, captopril, against HSV-1. By combining qualitative analysis of cellular structure with quantitative measurements of cellular viability and viral replication, and molecular modeling, we have demonstrated that captopril effectively decreases the cytopathic effects induced by HSV-1 by mediating viral replication. Due to the difficulty of treating viral infections, this work has the potential to be applicable across a broad spectrum of other viruses and raises additional questions regarding systemic effects of HSV-1 infection, such as hypertension and other cardiovascular diseases in general. This work provides a platform to further investigate the mechanisms of captopril, an already approved drug, as an effective therapeutic alternative for common HSV-1 infection.
Figshare: Quantitative and qualitative data assessing the antiviral effects of Captopril, an ACE-1 inhibitor, on HSV-1, https://doi.org/10.6084/m9.figshare.12881441.v122.
This project contains the underlying data files:
- Captopril Dose-Response data.xlsx [Captopril dose-response data]
- HSV-1 & Captopril Time Course Data.xlsx [HSV-1 and captopril time course data]
- JPG files - Qualitative comparison of cell morphology of different treatment groups
- Virus Replication Assay-GFP+ Cells.xlsx [HSV-1 viral replication quantitation based on GFP+ cells]
- PNG files - Qualitative evaluation of GFP+ cells in different treatment groups at different time points
- Captopril-HSV-1 Interaction (dragged).pdf [Molecular modelling: Captopril has a favorable interaction with HSV-1 gD]
- ICP0 gene expression data.csv [Captopril treatment down-regulates HSV-1 ICP0 expression]
Data are available under the terms of the Creative Commons Attribution 4.0 International license (CC-BY 4.0).
The authors acknowledge the kind support of the Fraser Laboratory (University of Pennsylvania), the Bertke Laboratory (Virginia Polytechnic Institute and State University), the Martinez Laboratory (Tuskegee University), and the Yates Laboratory (Tuskegee University).
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
No
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Partly
Are the conclusions drawn adequately supported by the results?
No
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Herpesvirus biology
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Partly
Are all the source data underlying the results available to ensure full reproducibility?
Partly
Are the conclusions drawn adequately supported by the results?
Partly
References
1. Di Giovine P, Settembre EC, Bhargava AK, Luftig MA, et al.: Structure of herpes simplex virus glycoprotein D bound to the human receptor nectin-1.PLoS Pathog. 2011; 7 (9): e1002277 PubMed Abstract | Publisher Full TextCompeting Interests: No competing interests were disclosed.
Reviewer Expertise: cell physiology, virology
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 11 Sep 20 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)