Keywords
Coelogyne kaliana, Coelogyne stenochila, Coelogyne tiomanensis, endemic species, RAPD, SCAR, species identification, taxon delimitation
Coelogyne kaliana, Coelogyne stenochila, Coelogyne tiomanensis, endemic species, RAPD, SCAR, species identification, taxon delimitation
1. We added Seidenfaden and Wood, 1992 into introduction
2. We replace Figure 1, showing the vegetative structure
3. We replace Table 2 that includes all the suggested information by reviewer 1.
4. We insert ". Thus, a comprehensive screening on more samples of the three species was not feasible in this case. Furthermore, the RAPD-SCAR markers have yet to be tested on other Malaysian Coelogyne to confirm the specificity of the markers. Hence, the RADP-SCAR markers should be tested on the more commonly available Coelogyne species such as C. asperata and C. speciosa using in future experiments." into discussion section as marked in 'track changes'
To read any peer review reports and author responses for this article, follow the "read" links in the Open Peer Review table.
The genus Coelogyne L. belongs to the Orchidaceae family, which is comprised of about 200 sympodial species distributed throughout India, southwest China, southeast Asia and the Fiji Islands. Their main centres of diversity are Borneo, Papua New Guinea, Sumatra and the Himalayan range (Butzin, 1992; Gravendeel et al., 2001). There are 26 species of Coelogyne in Peninsular Malaysia based on the latest Checklist of Orchids of Peninsular Malaysia (Ong et al., 2017) and Seidenfaden & Wood, 1992. Amongst them, Coelogyne kaliana, Coelogyne stenochila and Coelogyne tiomanensis are endemic to Peninsular Malaysia (Figure 1). They are mostly found in the highland regions at elevations of 1200 m above sea level. These three Peninsular Malaysian endemic species are very rare and are present in small populations. Endemic orchid species are national treasures, which have high commercial values among orchid collectors and enthusiasts. This leads to their illegal and indiscriminate collections from the wild. Moreover, the survival of many endemic species may be jeopardized due to rapid climate change, which causes their populations to decline gradually. This eventually would reduce the likelihood of them to being found in their natural habitats (Ong, 2013). Scientific and efficient discriminations of morphologically similar species have been indispensable in achieving the ultimate goal of preservation and conservation of endemic orchid species. Apart from their distinctive floral appearances, C. kaliana, C. stenochila and C. tiomanensis are morphologically similar with indistinguishable common vegetative structures. Thus, it is crucial that molecular markers be developed, which would enable us to easily distinguish among them taxonomically.

(A) Coelogyne tiomanensis; (B) Coelogyne kaliana and (C) Coelogyne stenochila (all with flower and vegetative structure).
Random Amplified Polymorphic DNA (RAPD) is a polymerase chain reaction (PCR)-based method (Williams et al., 1990) that uses short and arbitrary oligonucleotides with GC contents of at least 50% to produce amplification products at random. The short oligonucleotide primer of approximately 10 bp in length is called a decamer and serves as both the forward and reverse primers. The amplification process in RAPD analysis is performed on total genomic DNA. Thus, it can be used to study genetic polymorphisms within the whole genome instead of just polymorphisms within a single genetic region. It also allows the amplification of random fragments of the genome without prior sequence knowledge. The main constraint of RAPD is its low fidelity. However, the conversion of RAPD markers into new, longer and specific Sequence Characterized Amplified Region (SCAR) markers can significantly improve reliability and reproducibility. Some other PCR-based methods, such as single locus microsatellites, Inter-Simple Sequence Repeats (ISSR) and Amplified Fragment Length Polymorphisms (AFLP), show better reproducibility of amplifications than the RAPD method. But, SCAR marker developments from these methods are often costlier, time consuming and laborious (Kumar & Gurusubramanian, 2011) when compared to its development from the RAPD method.
The SCAR marker (Paran & Michelmore, 1993) is a robust and reliable method used to detect and differentiate different samples by using specific primers derived from RAPD, ISSR, AFLP and other DNA markers. SCAR markers can discriminate among closely related samples or species by amplifying products of different sizes or no amplification in non-targeted samples and positive amplifications in targeted samples. Over the past few years, this method had been widely adopted in the identification of morphologically similar but genetically different organisms. A large number of robust and reliable SCAR markers had been successfully developed through the RAPD-based method for Pisum sativum (Srivastava et al., 2012), Oryza sativa (Semsang et al., 2013), Sorghum halepense (Zhang et al., 2013) and even for orchids, such as Phalaenopsis (Goh et al., 2005; Niknejad et al., 2009) and Paphiopedilum (Sun et al., 2011). However, currently there is still no genetic marker for the species identification of Peninsular Malaysia’s endemic Coelogyne species.
Hence, in this present investigation, RAPD-based SCAR markers were developed to distinguish among the three endemic Coelogyne species in Peninsular Malaysia. RAPD primers were used to screen the DNA samples of these three species to amplify reproducible species-specific bands. A single or a combination of two SCAR markers, which we developed, allowed us to successfully distinguish among the three morphologically similar and closely related Peninsular Malaysian endemic Coelogyne species. To our best knowledge, this is the first study on developing species-specific SCAR markers to identify the three endemic Coelogyne orchids of Peninsular Malaysia.
Due to the rarity of these species, only two samples of C. kaliana (YKH 020, Genting Highlands and YKH 004, Cameron Highlands, Malaysia), one sample of C. stenochila (YKH 031, Gunung Tahan, Malaysia) and one sample of C. tiomanensis (FRI 75329, Tioman Island, Malaysia) were collected from their native locations in Peninsular Malaysia. A small piece (3 cm × 3 cm) of fresh leaf from each sample was used for the DNA extraction. Genomic DNA was extracted based on the conventional CTAB method (Doyle & Doyle, 1987). The DNA pellets were suspended in 50 μl sterile DNase free water and kept at −20 °C until ready for use. The purity and concentration of the DNA samples were determined using a spectrophotometer (NanoDrop, USA).
A total of 16 decamer universal RAPD primers (synthesized by First Base Laboratory, Serdang, Malaysia), as shown in Table 1, were used to amplify the DNA samples of the three Malaysian endemic Coelogyne species. These sequences were obtained from published articles (Table 1) where they have been used successfully in other orchid species. The RAPD amplification reactions were performed in a 10 μl volume containing 50 ng of plant DNA, 1.0 μM of RAPD primer (First Base Laboratory), 1 × PCR buffer (Promega, USA), 2.5 mM of MgCl2 (Promega, USA), 0.1mM dNTPs mix (Promega, USA), 0.5 units of Taq DNA polymerase (Promega, USA), using a thermal cycler machine (Eppendorf Master Cycler Gradient, Hamburg, Germany), The run conditions were: 1 cycle of initial denaturation at 95°C for 10 min; 45 cycles of denaturation at 95°C for 1 min, annealing at 35°C for 2 min and extension at 72°C for 2 min; and 1 cycle of final extension at 72°C for 10 min. The amplicons were electrophoresed on 1.2% TBE (Tris-borate-EDTA) agarose gel for 90 minutes at 70 V and stained with 0.5μg/mL ethidium bromide. The gel was visualised under a UV transilluminator (UVIdoc, USA) and the gel image was taken using a camera (UVItec, UK). Species-specific bands were identified from the bands produced during the RAPD amplifications.
| Primer code | Sequence 5'−3' | Reference |
|---|---|---|
| OPA 03 | AGTCAGCCAC | (Grünanger et al., 1998; Obara-Okeyo & Kako, 1998; Parab & Krishnan, 2008) |
| OPA 04 | AATCGGGCTG | (Parab & Krishnan, 2008) |
| OPA 09 | GGGTAACGCC | (Grünanger et al., 1998; Parab & Krishnan, 2008) |
| OPA 12 | TCGGCGATAG | (Grünanger et al., 1998; Parab & Krishnan, 2008) |
| OPA 14 | TCTGTGCTGG | (Grünanger et al., 1998; Parab & Krishnan, 2008) |
| OPF 02 | GAGGATCCCT | (Benner et al., 1995) |
| OPF 03 | CCTGATCACC | (Benner et al., 1995) |
| OPF 04 | GGTGATCAGG | (Benner et al., 1995) |
| OPF 05 | CCGAATTCCC | (Benner et al., 1995) |
| OPF 06 | GGGAATTCGG | (Benner et al., 1995) |
| OPU 03 | CTATGCCGAC | (Goh et al., 2005; Lim et al., 1999; Niknejad et al., 2009) |
| OPU 08 | GGCGAAGGTT | (Goh et al., 2005; Niknejad et al., 2009) |
| OPU 10 | ACCTCGGCAC | (Goh et al., 2005; Niknejad et al., 2009) |
| OPU 12 | TCACCAGCCA | (Goh et al., 2005; Niknejad et al., 2009) |
| OPU 13 | GGCTGGTTCC | Goh et al., 2005; Niknejad et al., 2009 |
| OPU 16 | CTGCGCTGGA | Goh et al., 2005; Niknejad et al., 2009 |
Primers that consistently generated species specific PCR profiles in all the DNA samples were selected for primer design. The amplified PCR products were cloned and sequenced using the service provided by First Base Laboratory. All species-specific RAPD-generated PCR amplicons were cloned into pJET1.2/blunt cloning vector by chemical transformation into the E. coli competent cells. The white colonies, which contained the recombinant DNA (plasmid), were picked from LB/ampicillin/X-gal plates and the recombinant DNA was isolated from the bacterial culture. The targeted inserts were sequenced using SP6 and T7 universal sequencing primers.
The DNA sequences obtained were first subjected to nucleotide similarity searches using the BLASTN function of the NCBI database to check for significant similarities of the sequences with any sequences in GenBank Species-specific SCAR marker primer pairs were then designed based on the DNA sequences obtained. Species-specific SCAR marker primer pairs of 20–24 bases in length were designed with high stringency using the program Primer3 (Rozen & Skaletsky, 2000). Then, the designed SCAR marker primer pairs were synthesised by First Base Laboratory. Primer specificities were validated by amplifying the SCAR markers using the DNA samples of the three endemic Coelogyne species and by the same reaction mixture as was described in the section RAPD-PCR Amplification. The PCR profiles used for the amplifications of the designed SCAR primer pairs were: initial denaturation at 95°C for 10 min, 35 cycles of denaturation at 95°C for 1 min, annealing at 51 to 53°C (depending on the SCAR primer pair) for 2 min, extension at 72°C for 2 min, and a final extension cycle at 72°C for 10 min. The amplified PCR products were sequenced by First Base Laboratory, Serdang, Malaysia to confirm sequence specificities.
In the preliminary screenings of 16 random decanucleotide RAPD primers, all primers were able to amplify genomic DNA samples of Coelogyne kaliana, Coelogyne stenochila and Coelogyne tiomanensis, to produce a variable number of bands of different sizes. However, out of the 16 universal RAPD primers screened, only two RAPD primers, namely OPU 08 and OPU 12 showed a high level of consistency, producing distinct and reproducible fingerprint patterns in all the DNA samples tested. Amplification by primer OPU 08 consistently produced a clear band between 1 kb and 1.5 kb (Figure 2), which was unique to C. stenochila but absent in C. kaliana and C. tiomanensis. Primer OPU 08 also consistently amplified an intense band of roughly 750 bp that was specific to C. tiomanensis only and not found in C. stenochila and C. kaliana. Primer OPU 12 consistently amplified a distinct fragment of about 500 bp (Figure 2), which was present in all DNA samples of C. kaliana, but which was not observed in the DNA samples of C. stenochila and C. tiomanensis.
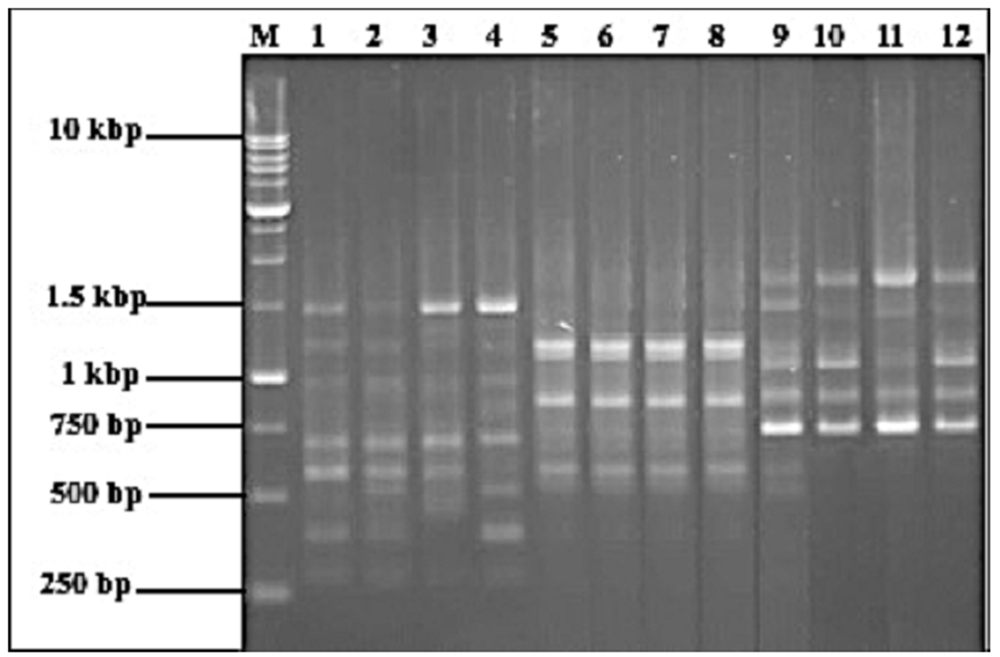
Lanes 1 and 2 are biological samples of C. kaliana (YKH004 and YKH020) while lanes 3 and 4 are technical replicates of YKH004 and YKH020 respectively; lane 5 is a biological sample of C. stenochila (YKH031) while lanes 6 to 8 are technical replicates of YKH031; lane 9 is a biological sample of C. tiomanensis (FRI75329) while lanes 10 to 12 are technical replicates of FRI75329. Lane M represents the 1.0 kb DNA ladder (Promega, USA). Fragments indicated by arrows were the specific bands, which were selected and subsequently sequenced for SCAR marker primer pair design.
Based on the RAPD fingerprinting results of OPU 08 and OPU 12, very few species-specific bands were observed. However, in selecting species-specific fragments for cloning and sequencing, other than focusing on bright and clear monomorphic band, fragment size was also taken into consideration. The ideal fragment size usually should range from 500 bp to 1.5 kbp, as too large a fragment faced difficulty in the cloning process; while too small a fragment provided less sequence for designing the SCAR marker primer pair. Therefore, a fragment of between 1 kbp to 1.5 kbp amplified by the OPU 08 primer was selected for C. stenochila, while another of approximately 750 bp was selected for C. tiomanensis. A fragment of about 500 bp amplified by primer OPU 12 was chosen for C. kaliana (Figure 3).

Lanes 1 and 2 are biological samples while lanes 3 and 4 are technical replicates of C. kaliana; lane 5 is a biological sample while lanes 6 to 8 are technical replicates of C. stenochila; lane 9 is a biological sample while lanes 10 to 12 are technical replicates of C. tiomanensis. Lane M represents the 1.0 kb DNA ladder (Promega, USA). Fragments indicated by arrows were the specific bands selected and sequenced for SCAR marker primer pair design.
Sequencing of the selected fragments yielded a sequence of 517 bp for C. kaliana, 1207 bp for C. stenochila and 742 bp for C. tiomanensis respectively. Homology searches using BLASTN showed that the fragments did not have similarity with any known nucleotide sequences in the NCBI database. The SCAR primer pairs of 20 to 24 bases each were designed with high stringency to ensure the specificity for each endemic Coelogyne species using these primer pair sequences. Details of the three designed primer pairs are shown in Table 2.
The efficacy and specificity of the designed SCAR primers were screened for using DNA samples of the three endemic Coelogyne species. SCAR primer pair CKL_f and CKL_r amplified a single, clear, distinct and easily identifiable band of 271 bp only in DNA samples of C. kaliana, while no amplification was observed for C. stenochila and C. tiomanensis (Figure 4). Hence, the SCAR primer pair CKL_f and CKL_r is a species-specific marker for C. kaliana.

Lane M represents the 1.0 kb DNA ladder (Promega, USA).
The SCAR primer pair CST_f and CST_r, designed based on the specific fragment selected from the RAPD profile of C. stenochila, was not species amplification specific as anticipated. However, this SCAR marker still allowed the differentiation of the three endemic Coelogyne species by amplifying a single and bright band of 854 bp in C. stenochila but two bands of different sizes (858bp and 372 bp) in C. tiomanensis, and no DNA amplification in C. kaliana (Figure 5).
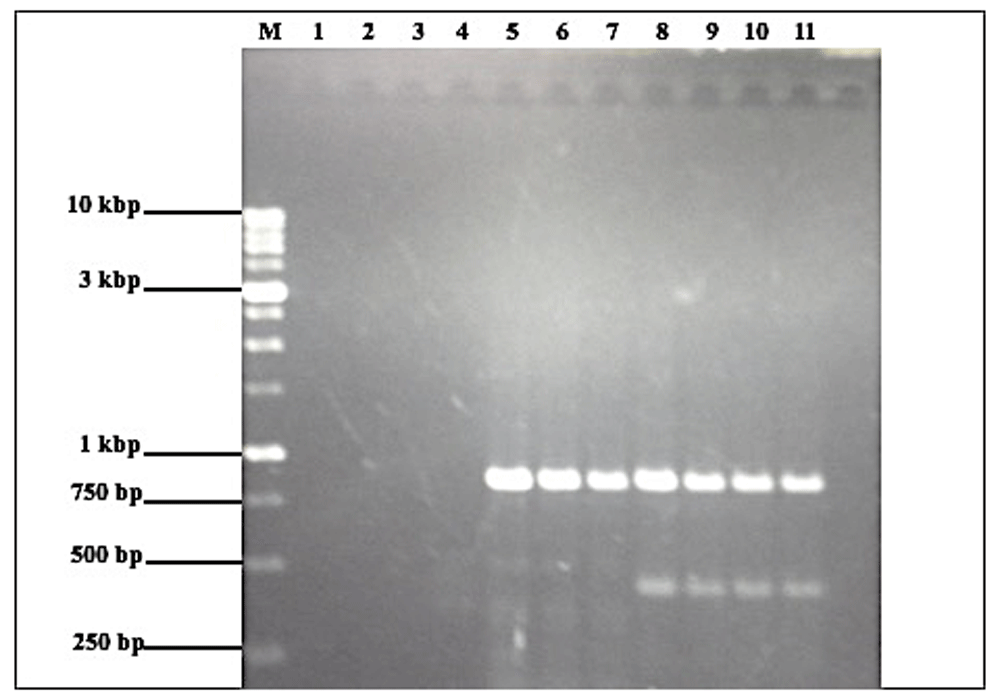
Lane M represents the 1.0 kb DNA ladder (Promega, USA).
The SCAR primer pair CTI_f and CTI_r, designed from the species-specific RAPD fragment for C. tiomanensis, was also not species amplification specific to C. tiomanensis. Nonetheless, it could still be used to differentiate C. kaliana from the other two endemic species, C. stenochila and C. tiomanensis. This SCAR marker amplified a single band of about 500 bp in both C. stenochila and C. tiomanensis but no DNA amplification in C. kaliana (Figure 6).
Based on the results of the present study, selected bands amplified by universal RAPD primers were successfully converted into specific SCAR markers. These SCAR markers were able to be used for the rapid identification and authentication of three Peninsular Malaysian endemic Coelogyne species, Coelogyne kaliana, Coelogyne stenochila and Coelogyne tiomanensis. Endemic species refers to species that are found exclusively or confined to a restricted geographical area. Both Coelogyne kaliana and Coelogyne tiomanensis were named after the regions from which they were first found. Coelogyne kaliana was first discovered in Gunung Ulu Kali of Genting Highlands in Peninsular Malaysia and was described by Cribb in 1982. Coelogyne tiomanensis, first described by Henderson in 1930, is found exclusively in Gunung Kajang of Tioman Island. Coelogyne stenochila, first described by Hook in 1890, can only be found in Pahang and Selangor states (Seidenfaden & Wood, 1992). The endemism and rarity of these Coelogyne species make their conservation of utmost importance. The accurate identification of these three morphologically similar species is one of the first steps towards achieving this goal by effectively preventing their illegal trading.
To the best of our knowledge, this is the first reported RAPD-SCAR study on the three endemic Peninsular Malaysian Coelogyne species. Although not all the SCAR markers developed in this study were species amplification specific, they could still elucidate the identity of the three endemic Coelogyne species efficiently and correctly. This was impossible through the traditional vegetative morphological characteristics method usually used by botanists. The main constraint in this study was that the number of samples available for use was very few due to the rarity and protected status of these endemic species. Thus, a comprehensive screening on more samples of the three species was not feasible in this case. Furthermore, the RAPD-SCAR markers have yet to be tested on other Malaysian Coelogyne to confirm the specificity of the markers. Hence, the RADP-SCAR markers should be tested on the more commonly available Coelogyne species such as C. asperata and C. speciosa in future experiments. Nonetheless, the SCAR markers developed in this study are new to science and are able to differentiate the three species accurately. This study further reinforced the robustness and suitability of SCAR makers as one of the leading methods for species identification of members of cryptic species complexes. This approach could also be used for other flagship species of economic and aesthetic importance. We hope that with the assistance of these markers, an exhaustive conservation plan could be developed in the immediate future to ensure the continued survival of these three rare endemic orchid species. Currently, the biggest threat to their survival is the high demand for them by orchid enthusiasts worldwide. This has resulted in them being obtained and traded illegally often by the sellers claiming that they are selling hybrid plants. It is expected that with the availability of the SCAR markers, which we have developed, this legal loophole can be successfully plugged.
Sequences are available from GenBank:
Coelogyne kaliana voucher YKH 004 maturase K (matK) gene, partial cds; chloroplast, Accession number MK398222.1: https://www.ncbi.nlm.nih.gov/nuccore/MK398222.1
Coelogyne kaliana voucher YKH 020 maturase K (matK) gene, partial cds; chloroplast, Accession number MK398221.1: https://www.ncbi.nlm.nih.gov/nuccore/MK398221.1
Coelogyne kaliana voucher YKH 004 ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit (rbcL) gene, partial cds; chloroplast, Accession number MK398165.1: https://www.ncbi.nlm.nih.gov/nuccore/MK398165.1
Coelogyne kaliana voucher YKH 020 ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit (rbcL) gene, partial cds; chloroplast, Accession number MK398164.1: https://www.ncbi.nlm.nih.gov/nuccore/MK398164.1
Coelogyne kaliana voucher YKH 004 tRNA-Leu (trnL) gene and trnL-trnF intergenic spacer, partial sequence; chloroplast, Accession number MK356248.1: https://www.ncbi.nlm.nih.gov/nuccore/MK356248.1
Coelogyne kaliana voucher YKH 004 small subunit ribosomal RNA gene, partial sequence; internal transcribed spacer 1, 5.8S ribosomal RNA gene, and internal transcribed spacer 2, complete sequence; and large subunit ribosomal RNA gene, partial sequence, Accession number MK356194.1: https://www.ncbi.nlm.nih.gov/nuccore/MK356194.1
Coelogyne kaliana voucher YKH 020 small subunit ribosomal RNA gene, partial sequence; internal transcribed spacer 1, 5.8S ribosomal RNA gene, and internal transcribed spacer 2, complete sequence; and large subunit ribosomal RNA gene, partial sequence, Accession number MK356193.1: https://www.ncbi.nlm.nih.gov/nuccore/MK356193.1
Coelogyne kaliana voucher YKH 020 tRNA-Leu (trnL) gene and trnL-trnF intergenic spacer, partial sequence; chloroplast, Accession number MK356247.1: https://www.ncbi.nlm.nih.gov/nuccore/MK356247.1
Coelogyne tiomanensis voucher FRI 75329 maturase K (matK) gene, partial cds; chloroplast, Accession number MK398239.1: https://www.ncbi.nlm.nih.gov/nuccore/MK398239.1
Coelogyne tiomanensis voucher FRI 75329 small subunit ribosomal RNA gene, partial sequence; internal transcribed spacer 1, 5.8S ribosomal RNA gene, and internal transcribed spacer 2, complete sequence; and large subunit ribosomal RNA gene, partial sequence, Accession number MK356154.1: https://www.ncbi.nlm.nih.gov/nuccore/MK356154.1
Coelogyne tiomanensis tRNA-Leu (trnL) gene and trnL-trnF intergenic spacer, partial sequence; chloroplast, Accession number MK356265.1: https://www.ncbi.nlm.nih.gov/nuccore/MK356265.1
Coelogyne tiomanensis voucher FRI 75329 ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit (rbcL) gene, partial cds; chloroplast, Accession number MK398182.1: https://www.ncbi.nlm.nih.gov/nuccore/MK398182.1
Coelogyne stenochila voucher YKH 031 ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit (rbcL) gene, partial cds; chloroplast, Accession number MK398181.1: https://www.ncbi.nlm.nih.gov/nuccore/MK398181.1
Coelogyne stenochila voucher YKH 031 small subunit ribosomal RNA gene, partial sequence; internal transcribed spacer 1, 5.8S ribosomal RNA gene, and internal transcribed spacer 2, complete sequence; and large subunit ribosomal RNA gene, partial sequence, Accession number MK356153.1: https://www.ncbi.nlm.nih.gov/nuccore/MK356153.1
Coelogyne stenochila voucher YKH 031 maturase K (matK) gene, partial cds; chloroplast, Accession number MK398238.1: https://www.ncbi.nlm.nih.gov/nuccore/MK398238.1
Coelogyne stenochila tRNA-Leu (trnL) gene and trnL-trnF intergenic spacer, partial sequence; chloroplast, Accession number MK356264.1: https://www.ncbi.nlm.nih.gov/nuccore/MK356264.1
Open Science Framework: Development of species-specific SCAR markers for identification and authentication of three rare Peninsular Malaysian endemic Coelogyne (Orchidaceae) orchids, http://doi.org/10.17605/OSF.IO/35XBG (Go, 2020) (project registered on September 1st, 2020; https://osf.io/dxtb2).
This project contains the following underlying data:
Sequences resulted from cloning.
Uncropped/unedited images of RAPD fingerprint blots for the 16 decamer universal RAPD primers.
Uncropped/unedited images of SCAR primer pair blots.
Data are available under the terms of the Creative Commons Attribution 4.0 International license (CC-BY 4.0).
The authors are also grateful to the Forest Research Institute Malaysia (FRIM) for providing the Coelogyne tiomanensis plant sample, and Forest Department of Peninsular Malaysia for giving us permission to access the study areas where the other samples were collected. Permit numbers are as follows: (a). National Park, Pulau Tioman - JPHL&TN(IP):80-4/2 Jld 17(11) dated 22 October 2013; (b) National Park, Gunung Tahan - JPHL&TN(IP):80-4/2 Jld 16 dated 3 June 2013.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Orchid biology, Horticulture
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Not applicable
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
References
1. G Seidenfaden, JJ Wood: The Orchids of Peninsular Malaysia and Singapore. 1992.Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Orchid biology
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Partly
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Plant Systematic, Plant Anatomy and Micromorphology, Palynology
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 2 (revision) 05 Jan 21 |
read | |
|
Version 1 21 Sep 20 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Thank you,
Rusea
Thank you,
Rusea
Thank you,
Rusea