Keywords
Nevirapine plasma concentration, pharmacogenetics and clinical parameters, HIV-1 patients in Kenya
Nevirapine plasma concentration, pharmacogenetics and clinical parameters, HIV-1 patients in Kenya
The use of antiretroviral therapy (ART) is an integral component in reducing the burden of HIV globally1. In 2018, it was estimated that 68% (826,000) of adults and 73% (71,500) of children in Kenya needing ART were actually receiving it1. A remarkable scale up of ART has put Kenya on track to reach the target of AIDS-related deaths1. Kenya is among the countries that have adopted the 2015 World Health Organization (WHO) recommendations, which require immediate initiation of ART to people diagnosed with HIV, aimed at increasing ART access further2. At the time of the study, the first-line ART guidelines for children, youth and adults in Kenya typically contained a backbone of two nucleoside reverse transcriptase inhibitors (NRTIs; zidovudine [AZT], stavudine [d4T], tenofovir [TDF] or lamivudine [3TC]), plus one non-nucleoside reverse transcriptase inhibitor (NNRTI): either nevirapine (NVP) or efavirenz (EFV)2.
NVP is a particularly extensively prescribed antiretroviral drug in developing countries mainly because of its efficacy, availability, low cost and use in prevention of vertical HIV transmission3. However, a higher incidence of rashes (which can be severe and life-threatening, such as Stevens-Johnson syndrome) associated with NVP4, the association with a rare but potentially life-threatening risk of hepatotoxicity5 and a low genetic barrier to resistance are the fundamental factors that restrict the use of NVP6. A study in Kenya reported that 35% of patients on NVP experienced treatment failure7. Factors such as unavailability of ART stock, poor prescribing practice and prompt antiretroviral refill are known to significantly affect treatment outcomes8.
Generally, the most important aspect of ART management is ideal drug exposure9; extensive use of, or suboptimal exposure to, ART, especially NNRTIs (NVP and EFZ), bestows countless risks to the success of treatment10. Studies in the USA, Europe and Asia have shown that low NNRTI concentrations are predictive of virologic failure11,12 and highly ART drug-resistant HIV quickly develops when administered in suboptimal regimens13. In Africa, studies have reported expansive person to person variation in plasma antiretroviral levels among patients with management failure, with a large percentage falling out of the therapeutic window14,15. Though countable reports have shown plasma NVP concentrations affect treatment outcome16, therapeutic drug level quantification is currently not part of HIV management in many countries, including Kenya15.
NVP is majorly metabolized by cytochrome P450 2B6 (CYP2B6) and minorly by CYP3A isoforms17. The occurrence of single-nucleotide polymorphisms (SNPs) especially on 516GT and 983TC, are correlated with significant changes in hepatic enzyme metabolic activities17. Other factors such as body weight, age, clinical outcome, gender, CD4 and HIV viral load nadir are also important determinants of NVP plasma levels8,18.
To build up data aimed at personalizing ART treatment in Kenya, this study assessed the relationship between CYP2B6 516G>T and 983T>C polymorphism, HIV drug resistant mutation, and other clinical parameters with NVP concentrations among HIV-1 patients receiving ART treatment in western and coastal Kenya.
This study was approved by the Kenya Medical Research Institutes’ (KEMRI) Scientific Review Unit (SERU) (KEMRI/SERU/CVR/002/3214) and NACOSTI (NACOSTI/P/19/11747/28173). Before recruitment in this study, all patients filled in written informed consent for study participation.
This was a cross-sectional study conducted between August 2018 to January 2020 and was part of an ongoing study designed to establish a cost-effective laboratory method to monitor antiretroviral adherence in HIV-1 infected individuals on treatment in Kenya. Patients were recruited in this study if they were: (i) HIV-1 infected adults (aged above 18 years), attending the two HIV treatment programs, (ii) willing to voluntarily give written informed consent, (iii) able to read either English or Kiswahili, (iii) be on ARV treatment for 12 months, (iv) be on NVP based 1st line ARV treatment regimen and (v) HIV patients with viral load results at month 12 of treatment. The patients were categorized as either failing (HIV viral load >1000 copies/mL) or responding (HIV viral load <1000 copies/mL) to treatment.
Using the case control sample size formula described by Lemeshow et al.19, and using the previous study of Ahoua et al.20, which showed that among HIV patients with virological failure at month 12, 12.8% of them developed sub-optimal drug level/drug resistance. In this study, we wished to have a 90% chance of detecting an odds ratio significantly different from 1 at the 5 % level. Considering an odds ratio of 2 as an important difference between the two groups, a total of 376 (188 cases and 188 controls) were to be recruited from each of the two sites. A two-stage sampling method was then used to select patients meeting the inclusion criteria from the two sites. First, the overall number of patients meeting the inclusion criteria was generated based on the laboratory records. A total of 272 patients from Kisumu (western Kenya) and 105 patients from Malindi (costal Kenya) were then selected based on probability proportionate to size. Second, a consecutive sampling technique was used to obtain consent from and recruit every patient meeting the inclusion criteria until the required sample size was achieved.
An exhaustive structured interview (including demographic data, clinical history, adherence, HIV stigma and medical history) was used to collect patient-related information (see Extended data)21. These interviews were conducted by medical clinician or doctor employed by the two organizations and who regularly attends to these patients. Each interview was conducted in a separated private room and lasted for about 45 minutes.
After the interviews, the patients were sent to the phlebotomy room where blood specimens were drawn by a trained phlebotomist into ethylenediaminetetraacetic acid (EDTA)-containing Vacutainers® (BD, US) for determination of NVP plasma concentration and CYP2B6 516G>T and 983T>C genetic analysis. From each of the patients, approximately 5ml of intravenous blood was collected in EDTA tubes. Blood samples were centrifuged at 20,000g to collect plasma, which was stored at -20°C at each research site until it was shipped in dry ice to the KEMRI Nairobi laboratory for storage and laboratory testing.
RNA was extracted manually from 1ml of all HIV-1 samples using QIAmp viral RNA mini kit (Cat. No. 52906, Qiagen Inc., USA) according to the manufacturer instructions. Purified RNA was eluted in 60μL of molecular grade water. A volume of 10μL of RNA extract was used for quantification with the Generic HIV Viral Load assay (Biocentric, Bandol-France). The cycling conditions comprised of 50°C for 10 minutes and 95°C for 5 minutes, followed by 50 cycles of 95°C for 15 seconds and 60°C for 1 minute. Amplification and data acquisition were carried out using the ABI Prism 7300 Sequence Detection System (Applied Biosystems) and the detection cut-off value was 60 HIV-1 RNA copies/ml.
The NVP plasma concentrations were measured using a Xevo TQ-S tandem quadrupole mass spectrometer (Waters Corporation, U.S.A) designed for ultra-high performance as described by Reddy et al.22. First, the HIV virus was inactivated as follows. Into a 1.5ml Eppendorf tube, 50μl of plasma and 5μl internal standard (2μg/ml nevirapine, purity: 100 %, from Vivan Life Sciences, Mumbai, India prepared in methanol) was added. This was heated at 65°C for 10 minutes, followed by 10 minutes cooling at room temperature. 100μl cold methanol (-20oC) was then added to each sample and kept at -20°C for 10 minutes. This was followed by eight minutes of centrifugation at 20,000g, 20°C to collect the supernatant in a clean 1.5ml tube. Then, 850μl ammonium acetate buffer (pH = 3.00) was added to the supernatant and briefly centrifuged. The sample was considered safe to be handled in a non P3 laboratory.
Solid phase extraction was carried out using Bond Elut C18 cartridges. The cartridges were prepared and placed onto the Visiprep Vacuum Manifold with standard lid (Merck, Germany). The Bond Elute C18 150×4.6mm, 5μm column was conditioned by first passing through 1ml methanol, followed by 1ml ultrapure water. Each column was then charged with 150μl samples containing 850μl ammonium acetate buffer (pH = 3.00), followed by twice cleaning using 1ml ultrapure water. The first cleaning was collected into clean separate tube while the second water cleaning collected in the waste tubes. The columns were vacuum dried (5–10 kPa). NVP elution was carried out at a flow rate of 1ml/min twice using 500μl methanol with vacuum drying between the two elutions. Elutes were then completely evaporated using Thermo Scientific™ Reacti-Vap™ Evaporators (Thermo Fisher Scientific Inc, USA) at 37°C for 30 minutes. This was then reconstituted using 100μl of equal parts 1:1 acetonitrile and water, vortexed briefly and transferred into 50ml capped vials and placed into the Xevo TQ-S for quantification. Approximately 1μl of the samples was injected automatically into the LC/MS/MS instrument and quantified within five minutes. NVP plasma concentration was categorized as <3100 ng/mL (below therapeutic range), 3100–4300 ng/mL (therapeutic range) and >4300 ng/mL (above therapeutic range) as previously defined23.
DNA was extracted from patients’ blood samples using the QIAamp DNA Blood Mini Kit (Cat. No. 51106, Qiagen GmbH, Hilden, Germany) according to the manufacturer’s instructions. Briefly, 20μl QIAGEN Protease and 200μl blood sample were added into a 1.5ml microcentrifuge tube and 200μl of Buffer AL added and mixed by pulse-vortexing for 15 seconds. This was incubated at 56°C for 10 minutes. This was followed by a series of washing and eventually the DNA was eluted using 200μl Buffer AE at 6000 × g (8000 rpm) for one minute. The quality of DNA was measured using a ND-1000 UV spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). Genotyping was carried out on an ABI 7500 Fast Sequence Detection System (Applied Biosystems, Foster City, CA, USA). SNPs were analyzed using the validated Taqman Genotyping Assays for CYP2B6 516G>T (rs3745274; assay ID C_7817765_60) and CYP2B6 983T>C (rs28399499; assay ID C_60732328_20), according to the manufacturer’s instructions. Briefly, in a final volume for each reaction of 25μl, consisting of 2x TaqMan Universal PCR Master Mix (Applied Biosystems, Foster City, CA, USA), 20x drug metabolizing genotype assay mix and 10ng genomic DNA. The PCR profile consisted of an initial step at 50°C for 2 minutes and 50 cycles at 95°C for 10 minutes and 92°C for 15 seconds. The plates were read using the allelic discrimination settings. The SNP assay was set up using SDS, version 1.3.0 as an absolute quantification assay. Post-assay analysis was done using SDS software. The results for CYP2B6 516G>T and 983T>C genotypes were defined as follows: homozygous wild type as 516GG or 983TT, heterozygous as 516GT or 983TC, and homozygous mutated as 516TT or 983CC.
The presence of HIV drug-resistant mutation was tested using an in-house genotypic method previously described Lehman et al.24. This involved the following steps:
RNA extraction. The viral RNA was extracted from plasma using QIAamp Viral RNA Extraction Kit (Cat. No. 52906, Qiagen Inc., USA) according to manufacturer’s instructions.
Nested PCR amplification and visualization. A nested PCR was then performed using AmpliTaq Gold (Roche Molecular Systems, Branchburg, NJ). Briefly, in the first round; HIV- 1 pol gene was amplified using primers (RT18: 5′ GGAAACCAAAAATGATAGGGGGAATTGGAGG 3′ and RT21: 5′ CTGTATTTCTGCTATTAAGTCTTTTGATGGG 3′) achieved as follows: one cycle of 45°C for one minute and 94°C for two minutes, followed by 35 cycles of 94°C for 30 seconds, 55°C for 30 seconds, and 72°C for one minute, with a final extension of 72°C for two minutes. The second-round amplification used primers (RT1: 5′ CCAAAAGTTAAACAATGGCCATTGACAGA 3′ and RT4: 5′ AGTTCATAACCCATCCAAAG 3′) was achieved as follows: one cycle of 94°C for two minutes and 30 cycles of 94°C for 30 seconds, 55°C for 30 seconds, and 72°C for one minute, with a final extension of 72°C for 10 minutes. The PCR amplification was confirmed by visualization with ethidium bromide staining of agarose gel. The nested reverse transcriptase-polymerase chain reaction (RT-PCR) amplified a positive sample of 645 base pairs.
BigDye sequencing reactions. The PCR positive samples were first cleaned to remove excess primers and nucleotides in a single step up using ExoSAP-IT™ PCR technology (Cat. No. 78200, Applied Biosystems, Foster City, CA, USA). Briefly, for each positive PCR product visualized on the gel, 5ul of the PCR product was mixed with 2ul ExoSAP-IT reagent and held at 37°C for 15 minutes, followed by 80°C for 15 minutes in a thermocycler.
The cleaned DNA was then amplified using a BigDye Terminator Kit (Cat. No. 4337457, Applied Biosystems, Foster City, CA, USA) and an ABI Prism 3300 Genetic Analyzer (Applied Biosystems, Foster City, US). Briefly, a 10µl reaction comprising 5.5µl DNA grade water, 2µl 5x BigDye buffer, 1ul BigDye 0.5µl primers (RT 1: 5’ CCAAAAGTTAAACAATGGCCATTGACAGA 3’ and RT4: 5’ AGTTCATAACCCATCCAAAG 3’), and 1ul PCR product template was amplified as follows: one cycle of 94°C for 10 seconds and 25 cycles of 94°C for 10 seconds, 50°C for 5 seconds, and 60°C for 4 minutes24.
This BigDye PCR amplification product was cleaned up using spin columns impregnated with Sephadex® G-50 (Sigma, US) and denatured using 10 ul Hi-Di™ Formamide and heated at 95°C for two minutes using a thermocycler. The 20ul of the denature BigDye product were sequenced using BigDye technology on an ABI 310 Genetic Analyzer (Applied Biosystems, Foster City, CA).
Drug resistant interpretation. The ART drug resistance mutations were identified using the Stanford University and International AIDS Society-USA website. Genotypic resistance was defined as the presence of resistance mutations associated with impaired drug susceptibility using the Stanford Genotypic Resistance Interpretation Algorithm.
Frequencies and percentages were used to present the sociodemographic data. The relationship between NVP plasma concentrations and CYP2B6 516G>T and 983T>C and the presence of HIV drug-resistant mutations were determined using Kruskal-Wallis test and Dunn’s test. Univariate and multivariate linear regression analyses were performed to determine the relationship between NVP plasma concentration and genetic polymorphisms, presence of HIV drug-resistant mutations and other clinical characteristics at the significance level of p<0.05. All statistical analyses were performed using STATA v 13 (StataCorp LP, Texas, USA).
The results from the 377 patients were assessed, of whom 272 (72.2%) were from Kisumu county (western Kenya), 223 (59.2%) were female and 114 (30.2%) had a HIV viral load of >1000 copies/mL25. The median age of the patients was 41 years (IQR = 34–49 years), with a median duration of living with HIV infection of five years (IQR = 1–11years) and a median duration since ART initiation of three years (IQR = 1–8 years). There were 306 (81.2%) patients currently taking lamivudine, nevirapine, tenofovir regimen, while 97 (25%) had missed an ART scheduled visit due to HIV-related illness and 205 (54.4%) reporting missing taking current ART at least once (Table 1).
The steady-state NVP plasma concentrations varied widely among patients, ranging from 4 ng/mL to 44,207 ng/mL (median 5179 ng/mL, IQR 2557–7453 ng/mL). Out of the total 377 patients, 96 (25.5%) had an NVP concentration <3100 ng/mL and 26 (6.9%) had an NVP concentration of 3100–4300 ng/mL, with the majority, 255 (67.6%), of the patients having an NVP concentration >4300 ng/mL (Table 2). No significant differences were found with regards to region of origin between patients with NVP levels <3100 ng/mL and those with NVP levels of 3100–4300 ng/mL or >4300 ng/mL (p=0.829). Further, no differences were observed with regards to gender, HIV RNA viral load, age and duration infected with HIV in patients with NVP levels >4300 ng/mL when compared to patients with lower NVP levels. Similarly, no significant correlations were observed with regards to age of sexual debut (p = 0.785), duration since ART initiation (p=0.888), initial ART regimen type (p=0.883), current ART regimen type (p=0.972), missing scheduled HIV care visit (p=0.644), or non-adherence to current ART regimen (p=0.769) in patients with NVP levels >4300 ng/mL when compared to patients with lower NVP levels (Table 1).
The number of patients with GG, GT and TT genotypes for the CYP2B6 516G>T SNP were 142 (37.7%), 187 (49.6%) and 48 (12.7%), respectively. In the case of the CYP2B6 983T>C SNP, the majority of patients (n = 326, 86.5%) had the homozygous wild type TT genotype. There were 48 patients (12.7%) who had the heterozygous mutant TC genotype, while three (0.7%) patients had the homozygous mutant CC genotype.
In this study, CYP2B6 516G>T and 983T>C SNPs were correlated with increased mean NVP plasma concentrations (Table 2). For CYP2B6 516G>T, patients who had the homozygous mutation (CYP2B6 516TT) had higher median NVP plasma levels (6753.5 ng/mL, IQR 4595.5–11046 ng/mL), as did those who were heterozygous for the mutation (CYP2B6 516GT; 5579 ng/mL, IQR 2960–8323 ng/mL), compared to those with the wild-type (CYP2B6 516GG; 3920.5 ng/mL, IQR 1416–6278 ng/mL) (p<0.0001). In the case of CYP2B6 983T>C, although the median NVP plasma concentrations were higher among patients who had the heterozygous genotype (CYP2B6 983TC; 5987 ng/mL, IQR 3101.5–9143 ng\mL) than those who had homozygous wild-type (CYP2B6 983TT; 5132 ng/mL, IQR 2394–7384 ng\mL), the distribution was not significant (p=0.33).
With regard to the presence of a HIV drug-resistant mutation, although there were only 31 (8.2%) patients with an NVP-based resistant mutation as opposed to 346 (91.8%) without, the median plasma NVP levels for those with resistant mutations was almost half (2283 ng/mL, IQR 18–5283 ng/mL) that of those without (5498 ng/mL, IQR 2960–7690 ng/mL) (p=0.001) (Table 2). The associations between log10-transformed plasma NVP concentrations and CYP2B6 genotypes and the presence of HIV drug-resistant mutations are shown in Figure 1.
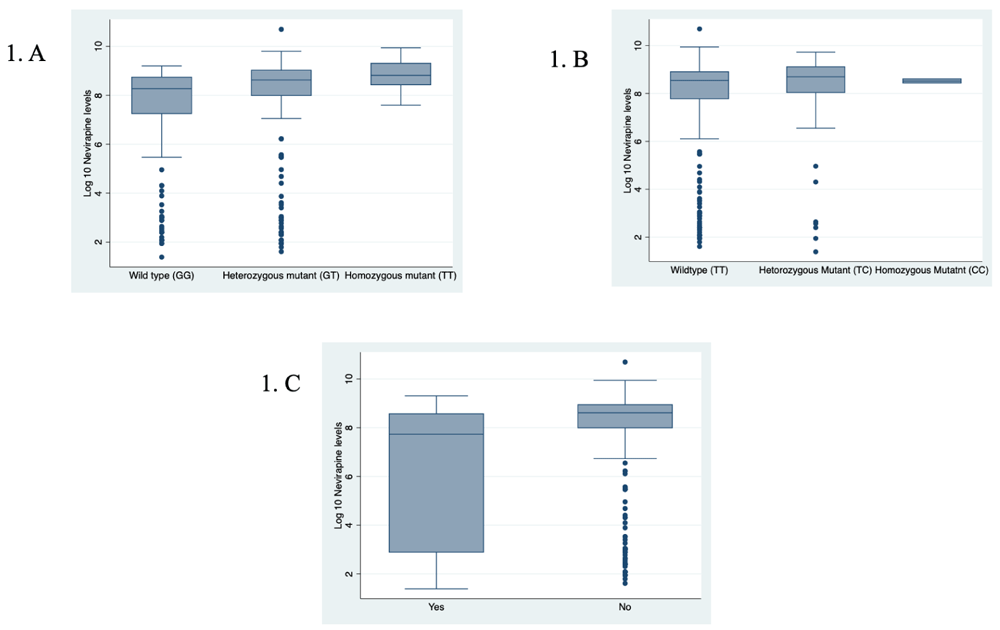
A) Log10-transformed nevirapine plasma levels for each CYP2B6 516G>T genotype: GG, GT, and TT. B) Log10-transformed nevirapine plasma levels for each CYP2B6 983T>C genotype: TC, TT and CC. C) Log10-transformed plasma NVP levels for the presence (yes) and absence (no) of HIV drug-resistant mutations.
In the multivariate linear regression model, factors that remained significantly associated with a higher NVP plasma levels included: the T allele of CYP2B6 G516T genotype (adjusted β 0.71, 95% CI 0.4–0.98; p<0.0001), male gender (adjusted β 0.45, 95% CI 0.01–0.9; p=0.047) and presence of drug-resistant virus (adjusted β 1.98, 95% CI 1.24–2.72; p<0.001) (Table 3).
In this study, in the two regions of Kenya reporting highest prevalence of HIV infection, we found wide variation in person-to-person NVP concentrations, ranging from 4ng/mL to 44,207 ng/mL (median 5179 ng/mL, IQR 2557–7453 ng/mL). This range is wider compared to other studies reported in Kenya26, in South Africa27 and in India23.
The therapeutic range of NVP plasma concentration is indicated as 3000 to 8000 ng/mL. In our study, the number of participants with plasma NVP levels within this therapeutic range was 255 (67.6%). The NVP plasma concentration required to achieve virologic control is indicated as >3000 ng/mL28. Reports associate NVP plasma concentrations lower than this threshold with poor virologic control and a concomitant increase in the number of NNRTI–resistant mutations29. The current study reports that 25.5% of patients had NVP plasma levels of <3100 ng/ml, associated with poor viral suppression. This prevalence was higher compared to those reported earlier in Kenya26 and in Italy30. This finding may be anticipated among patients with a stable and longer NVP treatment period, with 25% of them reporting non-adherence to medication.
The prevalence of CYP2B6 516T (37.7% GG, 49.6% GT and 12.7% TT) and CYP2B6 983C (86.5% TT, 12.7% TC and 0.7% CC) genotypes among these patients was comparable to those described among other African ethnic groups26,31. Studies have already established the importance of CYP2B6 gene polymorphisms as a key determinant influencing NVP plasma concentrations32,33. In the current study, patients who had both CYP2B6 516G>T and CYP2B6 983T>T genotypes had higher plasma NVP concentrations. This effect was more pronounced among patients who were homozygous for the mutant (CYP2B6 516 TT). Even though this study was cross-sectional, several longitudinal studies have reported a genotype dose-dependent increase in NVP plasma concentration among carriers of the CYP2B6 c.516 T allele26,30,34. For CYP2B6 983T>C, even though there were only 48 and three participants with the heterozygous mutant genotype (CYP2B6 983TC) and the homozygous mutant (CYP2B6 983CC), respectively, these genotypes were marked with raised NVP plasma concentrations. In agreement with previous work in Kenya26, which reported that the heterozygous TC genotype was associated with 55% higher NVP levels compared to individuals who had the wild type TT genotype. The SNP in CYP2B6 983T>C results in the variant protein CYP2B6*18 with I328T as the only amino acid change35. In vitro experiments have shown no measurable protein or activity with the expression of CYP2B6 983TC and is thus termed as a null allele36. Therefore, in other studies, the null status of CYP2B6 983TC has been associated with a greater impact compared to the CYP2B6 516GT26,34.
In this study, although there were only 8.2% patients with a HIV drug-resistant mutation to both NNRTIs and NRTIs, compared to 91.8% with susceptible viruses, median plasma NVP levels were almost half that of those who did not have drug-resistant HIV. In agreement with other reports, the presence of a HIV drug-resistant mutation is associated with higher NVP plasma concentrations37. Sustained optimal NVP plasma concentration is vital, given that only a single point mutation at specific position on the HIV-1 pol gene is associated with increased NVP resistance38. Development of HIV drug-resistant strains are shown to occur at the trough plasma NVP concentration of ≤3μg/ml, which has been correlated with increased risk of treatment failure18,39.
Beyond genetic background and viral dynamics, other patient pharmacoecological factors influencing the day-to-day concentration of drugs were evaluated. Although not significant, male patients had slightly higher NVP plasma concentrations compared to females, contrary to a report from South Africa40, which showed that females had higher median NVP levels than males. This difference in NVP plasma levels between genders has been attributed to the differences in body size and drug clearance rates between males and females. Older patients had higher median NVP plasma levels than younger ones, although this was not significant. This corresponds to studies that indicated more rapid NVP metabolism among younger children aged ≤8 years, who may require higher doses of NVP to achieve the therapeutic concentration23,41.
The importance of ART regimen type, adherence, and duration on ART for the success of HIV treatment and care, although not significant in the current study, has been widely studied9,42. Generally, unstable steady-state levels of NVP have been attributed to poor adherence9,42.
Our study had some limitations worth pointing out. First, due to limited resources available, the study could not recruit a larger number of patients. Second, the use of an NVP-based ART regimen in Kenya and other countries, especially developed countries, has been considerably reduced in the recent past, meaning that this study could be relevant to a restricted number of patients. Third, this study assessed well-known SNPs (CYP2B6 516T and CYP2B6 983C), whose relationship with NVP metabolism has been established. Other SNPs or other regulatory genes could also be important in NVP metabolism. Fourth, the outcomes from one population may not be generalized to other populations due to differences in drug effects between those of different ethnicities and body weights. Fifth, this was a cross-sectional study, which only permitted the description of the relationship between NVP plasma concentrations, patient genetics and a few pharmacoecologic factors and not a causal conclusion. Such outcomes can be confirmed in a longitudinal study.
These limitations notwithstanding, our study, conducted in two regions of Kenya marked by high prevalence of HIV infection, shows the significant role of CYP2B6 polymorphisms in NVP plasma concentrations. NVP plasma concentration is linked to the emergence of NVP-related resistant mutations. Moreover, patient pharmacoecologic factors, such as gender, age and ART adherence, are key in influencing NVP plasma concentration. With the consistent advancement of technical know-how, even in resource-limited settings like Kenya, individualization of ART is today reachable with the use of pharmacogenomics.
Figshare: Role of pharmacogenetics and clinical parameters on nevirapine plasma concentration among HIV-1 patients receiving antiretroviral therapy in Kenya. https://doi.org/10.6084/m9.figshare.11977680.v125
Figshare: Role of pharmacogenetics and clinical parameters on nevirapine plasma concentration among HIV-1 patients receiving antiretroviral therapy in Kenya. https://doi.org/10.6084/m9.figshare.1203369921
This project contains the following extended data:
Data are available under the terms of the Creative Commons Zero "No rights reserved" data waiver (CC0 1.0 Public domain dedication).
We wish to acknowledge the study patients, all the staff of the respective comprehensive care clinics in the two regions of Kenya. The authors thank the late Brian Khasimwa and Erickson Angira for HIV drug resistant genotyping and drug plasma measurement.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Partly
Are all the source data underlying the results available to ensure full reproducibility?
Partly
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: HIV, SARS-CoV-2
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
No
If applicable, is the statistical analysis and its interpretation appropriate?
No
Are all the source data underlying the results available to ensure full reproducibility?
Partly
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: HIV, Infectious Diseases, SARS-CoV-2.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 14 May 20 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)