Keywords
Pseudomonas, Antibiotics Resistance, bacterial pathogen, P. aeruginosa
This article is included in the Pathogens gateway.
This article is included in the Antimicrobial Resistance collection.
Pseudomonas, Antibiotics Resistance, bacterial pathogen, P. aeruginosa
The emergence of Gram-negative bacterial antibiotics resistance is a growing threat to antibiotic therapy. The bacterial pathogens in the genus Pseudomonas are mostly opportunistic that cause great damage and loss of life (Breidenstein et al., 2011; Evans et al., 2008; Kawai, 1974; Lalucat et al., 2006). These Pseudomonas pathogens are pervasive that is able to infect, survive and proliferate in a wide range of biotic and abiotic environments (Azam & Khan, 2019; Silby et al., 2011). Presently, there are seven groups in the genus Pseudomonas, with P. aeruginosa being the most pathogenic (Barbier et al., 2013) and which causes high morbidity and mortality in cystic fibrosis patients and immunocompromised individuals (Sadikot et al., 2005). The other Pseudomonas pathogenic groups include P. fluorescens (Biaggini et al., 2015), P. putida (Fernández et al., 2015), P. stutzeri (Lalucat et al., 2006) and P. syringae (Xin et al., 2018). P. aeruginosa is the major cause of infections in developed countries due to its highly evolved resistance to a wide variety of antibiotics (Hancock & Speert, 2000) making it very difficult to treat and limiting therapeutics (Breidenstein et al., 2011). Even though P. aeruginosa is the most studied, there are some other Pseudomonas species that exhibits opportunistic pathogenic behavior to animals (P. fluorescens, P. putida and P. stutzeri) (Azam & Khan, 2019) and plants (P. syringae) (Xin et al., 2018). Most of the known antimicrobial resistance (AMR) gene and mechanistic studies are focused on P. aeruginosa with little attention directed to other Pseudomonas pathogens. Hence, there is need to investigate different Pseudomonas pathogens genomes for diverse antibiotic resistance genes (ARGs) and mechanism. Therefore, this brief report concisely revealed the ARGs, mechanisms and drugs in P. aeruginosa in comparison to other Pseudomonas pathogens.
The complete genomes of the five groups of Pseudomonas pathogens, which included the P. aeruginosa (NC_002516.2), P. fluorescens (NC_016830.1), P. putida (NC_002947.4), P. stutzeri (NC_015740.1) and P. syringae (NC_007005.1) fasta file sequences, were downloaded from The National Center for Biotechnology Information Genome database. The five Pseudomonas pathogen genomes were selected to represent the Pseudomonas groups. The fasta file format of the genome sequence of bacteria were thoroughly analyzed for ARGs on the bulk analysis Resistance Gene Identifier (RGI) 5.1.0, CARD 3.0.9 Platform (Alcock et al., 2020) to extract AMR Genes, AMR Gene Family, Drug Class and Resistance Mechanism data. Default select criteria, which identified gene based on strict or perfect only was used. On the RGI platform, each genome sequence file was uploaded and all settings were left at default. The resistance genes, mechanism and drugs obtained from RGI platform were further analyzed using Prism 8 for number of ARG hits, gene family, mechanism and drug class per Pseudomonas species.
The five Pseudomonas pathogens show significant genome similarity (Table 1). P. stutzeri and P. putida, which have been reported to be opportunist pathogens to humans, were shown to be the closest relations to P. aeruginosa, with an average nucleotide identity (%) of 80.30 and 79.52. P. syringae, which infects plants, had the lowest average nucleotide identity (%) of 78.67. A number of antimicrobial resistance hits and genes were identified across the five Pseudomonas pathogens, but the highest hits were seen in the P. aeruginosa genome. The pathogen genome with the least hit was P. stutzeri (Figure 1A, B). The four major antibiotic resistance mechanisms identified were the efflux, inactivation, target alteration and efflux::target alterations in the P. aeruginosa genome. The efflux, inactivation and efflux::target alterations were identified in the P. fluorescens genome, while only the efflux and inactivation alterations were identified in the P. putida, P. stutzeri and P. syringae genomes (Figure 2A). The number of drug classes that P. aeruginosa was shown to be resistant to is also shown in Figure 2B.
| S/N | Pseudomonas groups | Pathogenic | Genome accession No. | Average nucleotide identity (%) |
|---|---|---|---|---|
| 1 | P. aeruginosa | Pathogenic to plants and animals (Azam & Khan, 2019) | NC_002516.2 | RG |
| 2 | P. fluorescens | opportunistic human pathogens (Biaggini et al., 2015) | NC_016830.1 | 79.25 |
| 3 | P. putida | opportunistic human pathogens (Fernández et al., 2015) | NC_002947.4 | 79.52 |
| 4 | P. stutzeri | opportunistic human pathogens (Lalucat et al., 2006) | NC_015740.1 | 80.30 |
| 5 | P. syringae | Pathogenic to plants (Xin et al., 2018) | NC_007005.1 | 78.67 |
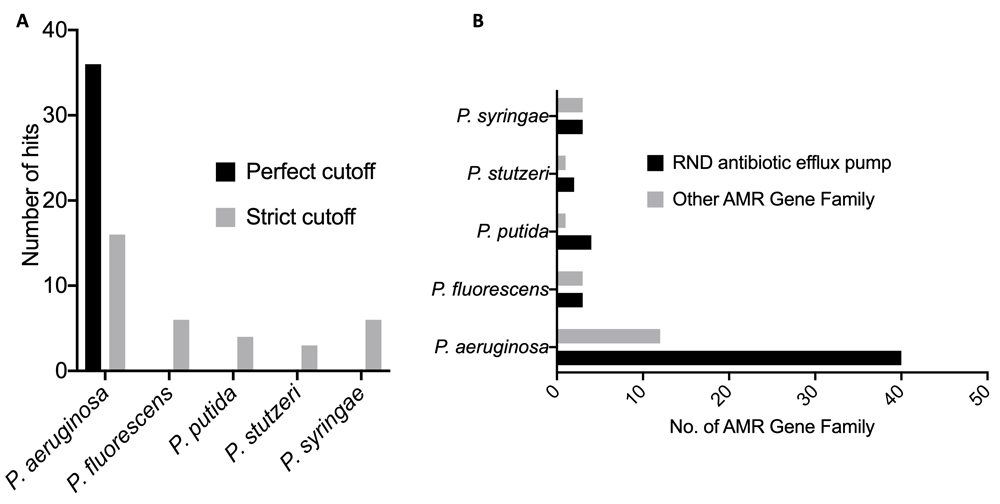
(A) The number of hit per each Pseudomonas pathogen genome (B). Number of ARGs per each Pseudomonas pathogen genome.
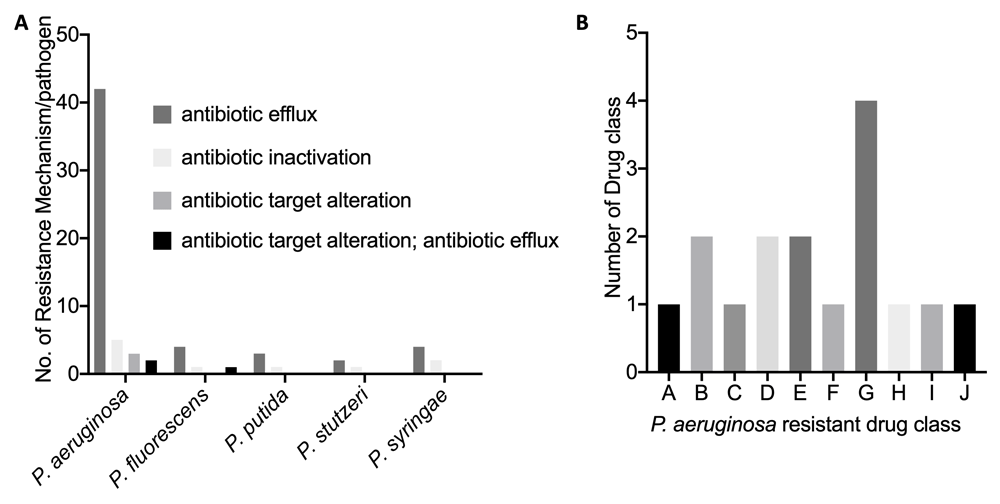
(A) Number of resistance mechanism per each Pseudomonas pathogen genome. (B) P. aeruginosa resistance drug class. Drug class key: A= aminoglycoside antibiotic; B= fluoroquinolone antibiotic, diaminopyrimidine antibiotic, phenicol antibiotic; C= macrolide antibiotic, carbapenem, tetracycline antibiotic, acridine dye, diaminopyrimidine antibiotic, phenicol antibiotic; D= macrolide antibiotic, fluoroquinolone antibiotic, aminoglycoside antibiotic, carbapenem, cephalosporin, cephamycin, penam, tetracycline antibiotic, acridine dye, phenicol antibiotic; E= macrolide antibiotic, fluoroquinolone antibiotic, aminoglycoside antibiotic, cephalosporin, penam, tetracycline antibiotic, aminocoumarin antibiotic, diaminopyrimidine antibiotic, phenicol antibiotic; F= macrolide antibiotic, fluoroquinolone antibiotic, cephalosporin, penam, tetracycline antibiotic, aminocoumarin antibiotic, diaminopyrimidine antibiotic, phenicol antibiotic; G= macrolide antibiotic, fluoroquinolone antibiotic, monobactam, carbapenem, cephalosporin, cephamycin, penam, tetracycline antibiotic, peptide antibiotic, aminocoumarin antibiotic, diaminopyrimidine antibiotic, sulfonamide antibiotic, phenicol antibiotic, penem; H= peptide antibiotic; I = phenicol antibiotic; J = sulfonamide antibiotic.
There is an increasing interest and focus in the antibiotic resistance in pathogenic Pseudomonas (Blair et al., 2014; Blanco et al., 2016; Li et al., 2015; Soto, 2013; Webber & Piddock, 2003). Hence, this report took advantage of the available Pseudomonas pathogens genomes and analysed for antibiotic resistance genes and mechanisms against different available drugs.
The significant number of ARGs and mechanisms were identified in the genome of P. aeruginosa, which is more virulent and well-studied compared to other species. P. aeruginosa also infects a wide range of plants and animals, including humans (Azam & Khan, 2019; Hancock & Speert, 2000; Sadikot et al., 2005). P. aeruginosa is of great medical importance due to its exhibition of multidrug resistance and its association with serious illnesses (Breidenstein et al., 2011; Evans et al., 2008; Gonzalez et al., 2019).
The most common resistance mechanism of Pseudomonas pathogens is the antibiotic efflux pump mechanism. Although this mechanism is most often seen in P. aeruginosa, it is also found in other Pseudomonas pathogen genomes. It has long been known that the antibiotic efflux pump is a key mechanism of resistance in Gram-negative bacterial pathogens (Blair et al., 2014; Blanco et al., 2016; Soto, 2013; Webber & Piddock, 2003). An antibiotic resistance strain’s efflux pumps allow it to regulate itself by excluding toxic substances, including antimicrobial drugs (Blair et al., 2014; Li et al., 2015; Soto, 2013; Webber & Piddock, 2003).
The different ARGs and mechanisms against known drugs in P. aeruginosa in comparison to other Pseudomonas pathogen were investigated and concisely reported in this brief report. The findings in this report could be useful in understanding the use of chemotherapeutics against antibiotic-resistant strains of Pseudomonas pathogens.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Not applicable
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Antimicrobial resistance evolution
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
No
If applicable, is the statistical analysis and its interpretation appropriate?
Not applicable
Are all the source data underlying the results available to ensure full reproducibility?
No source data required
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 04 Aug 20 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)