Keywords
absorbance scanning, tumor microenvironment, compositions, endoscopic biopsy, colorectal cancer
This article is included in the Oncology gateway.
absorbance scanning, tumor microenvironment, compositions, endoscopic biopsy, colorectal cancer
The tumor microenvironment is an area surrounding a tumor consisting of various cells including stromal cells (Wang et al., 2017), immune cells (Winkler et al., 2020), endothelial cells, blood, and lymph vessels (Wei et al., 2020). The intra-tumoral composition of the tumor microenvironment can influence the tumor progression (Whiteside, 2008) and the clinical outcome of patients. To be able to determine the intra-tumoural composition, one common method is histopathological evaluation of the patient's biopsy (Schillaci et al., 2019). However, the biochemical composition of the biopsy such as metabolites and cytokines requires further tests such as immunohistochemistry of cell markers. (Oudijk et al., 2015) On the other hand, tumor progression may be influenced by chemical compositions within the tumor microenvironment such as cytokines and growth factors (Landskron et al., 2014; Yuan et al., 2016).
One of the simple methods that can be used to distinguish chemical compositions is spectrometry and potential differences of chemical compositions in malignancy such as changes in amino acid concentrations (Balan et al., 2019). Previously, Nogueira et al., (2021) showed that a tumor can be detected using diffuse reflectance spectroscopy in colorectal cancer diagnosis. Another study by Yang et al., (2016) showed the potentiality of visible-absorption spectroscopy as a biomarker to predict treatment and prognosis of esophageal cancer. In this study, we report absorbance scanning to distinguish three biopsies from three different patients. The absorbance scanning was carried out at a wavelength of 400–1000 nm using a spectrophotometer followed by Principal Component Analysis Orange Data Mining.
All subjects gave their written informed consent for inclusion before participating in the study. The study was conducted in accordance with the Declaration of Helsinki, and the protocol was approved by the Ethical Committee of the Faculty of Medicine, Universitas Indonesia (Protocol ID: 20-04-0643, version 02, June 29th, 2020). The tissue samples taken from an endoscopic biopsy from patients from Cipto Mangunkusumo Hospital, Jakarta were confirmed as cancer by an anatomic pathologist in the Department of Pathological Anatomy, Faculty of Medicine, Universitas Indonesia. They were then put into transport media in phosphate buffer saline (PBS) and transferred to the laboratory of the Indonesia Medical Education and Research Institute, Faculty of Medicine, Universitas Indonesia in less than two hours.
The samples were minced into smaller tissue fragments, transferred to 1.5 ml centrifuge tubes, and homogenized by TissueLyser II (Qiagen, Germany) for 10 minutes. The suspensions were diluted in PBS to 1 mL.
The samples underwent wavelength scanning at λ = 400–1000 nm using the Agilent Cary 60 UV-Vis spectrophotometer (Agilent, Unites States). The absorption spectrum of the lysate cell was examined and feature selection was carried out using principal component analysis (PCA) to simplify the wavelength dimensions into principal component (PC) features that were not correlated (Abdullah et al., 2021). The resulting PC features were then used to classify the cancer patient samples into a scatterplot. The absorption/absorption spectrum pre-processing and PCA analysis were performed using Orange Data-Mining version 3.28.0 (Orange Data Mining, RRID: SCR_019811) (Demšar et al., 2013).
In this study, we recruited three different patients namely CRC006, CRC009, and CRC011. In the samples collected from patients, CRC006 and CRC011 similar clinical characteristics such as histopathology results were observed. Welldifferentiated rectal adenocarcinoma with metastatic sites were found in both patients. The sample collected from patient CRC009 had different clinical characteristics compared to CRC006 and CRC011, and CRC009 was found to have undifferentiated adenocarcinoma without metastasis. Sample data are shown in Table 1. (Abdullah et al., 2021)
| Sample ID | Histopathology results | Metastasis |
|---|---|---|
| CRC006 | Well differentiated rectal adenocarcinoma | Yes |
| CRC009 | Undifferentiated adenocarcinoma | No |
| CRC011 | Well differentiated adenocarcinoma | Yes |
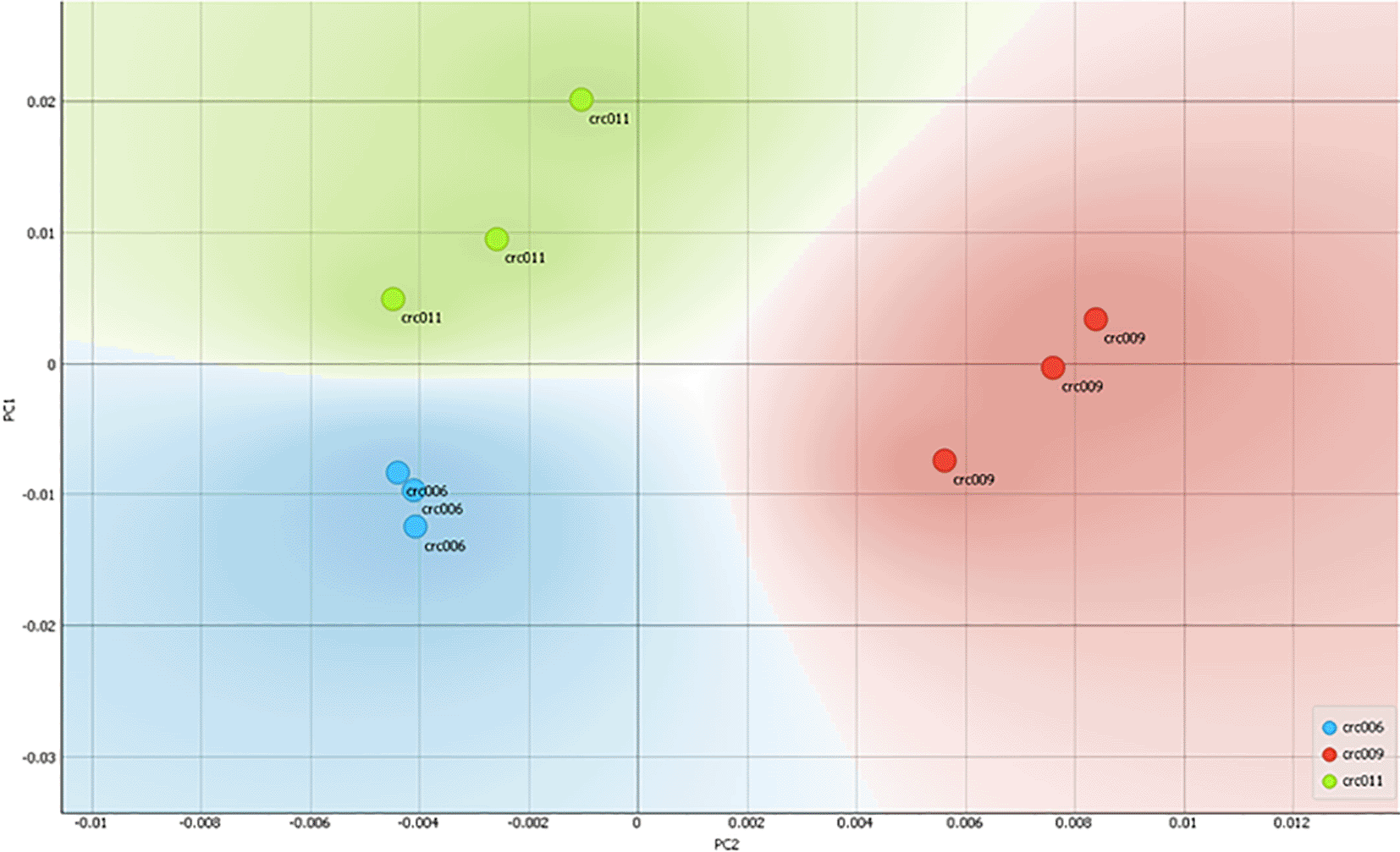
Figure 1 shows that each cell produces an absorption spectrum fingerprint in the wavelength range of 400–1000 nm. Just by observing the form of the absorption/absorption spectrum, it can be seen at first glance that fingerprints were only contributed to by the concentration of the molecule. To investigate the characteristic features of the spectrum, a principal component analysis (PCA) was carried out, which resulted in PC1 as the main feature (Abdullah et al., 2021). The results of the scatterplot with PC1 and PC2 showed that the sample CRC011 was well grouped in quadrant 1, CRC006 in quadrant 4, while CRC009 was spread between quadrants 2 and 3. From Figure 1 it can also be seen that each type of sample group appears to experience separation/classification when compared to other groups.
The light absorption spectrum is known as a “molecular fingerprint” which allows the analysis of molecular types and their amounts/concentrations. In Table 1, we show three representative colorectal cancer biopsies collected endoscopically from patients: CRC006 was a patient with welldifferentiated rectal adenocarcinoma with metastasis, CRC009 was from a patient with undifferentiated adenocarcinoma, CRC011 was from a patient with welldifferentiated adenocarcinoma with metastasis. Interestingly, based on the PCA analysis of the absorption spectrum of the lysate sample (shown in Figure 2), the CRC006 quadrant was closer to CRC011, which has similar clinical features of welldifferentiated adenocarcinoma and metastasis. On the other hand, CRC009 with undifferentiated and located far from CRC006 and CRC011 respectively, which indicated their closer chemical composition. Such existence of grouping indicates that there were differences in cellular composition between the three samples; although further studies with a larger number of samples are needed to further confirm the clustering that occurs.
In biological tissue samples, several molecules contribute to the absorption spectrum in the ultraviolet (UV) to nearinfrared (NIR) regions. Protein and DNA absorb in the UV region, whereas tryptophan, NAD+ (nicotinamide adenine dinucleotide), collagen, elastin, NADH, and FAD (flavin adenine dinucleotide) are important endogenous chromophores in biological tissue (Yang et al., 2016). FAD, tryptophan, and NADPH (nicotinamide adenine dinucleotide phosphate) have been used as a measuring tool to see the mitochondrial metabolic response before and after therapy such as in prostate cancer (Alam et al., 2017). In cell lysate samples such as in this study, further molecular analysis is needed to determine the differences in the composition of compounds in the absorption/absorption spectrum between lysates from cancer patients.
The combinations of principal component analysis (PCA) and absorbance scanning were able to group patients with similar histopathology and metastasis based on their chemical compositions. Although further investigation is needed, molecular fingerprinting using absorbance scanning to predict intratumoral similarity and difference is exciting.
Open Science Framework: Underlying data for ‘Potential use of absorbance scanning in differentiating tumor microenvironment compositions from endoscopic biopsy of colorectal cancer patients available at https://doi.org/10.17605/OSF.IO/QV7H5 (Abdullah et al., 2021).
This project contains the following underlying data:
Data are available under the terms of the Creative Commons Attribution 4.0 International license (CC-BY 4.0).
Written informed consent for publication of the patients’ details was obtained from the patients.
We thank HIBAH PUTI Q2 Universitas Indonesia (NKB4127/UN2.RST/HKP.05.00/2020) for funding our study.
We thank Dr Aryo Tedjo from the Department of Medical Chemistry and Drug Development Cluster for supporting the absorbance scanning and Orange mining data analysis.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
No
Is the study design appropriate and is the work technically sound?
No
Are sufficient details of methods and analysis provided to allow replication by others?
No
If applicable, is the statistical analysis and its interpretation appropriate?
Not applicable
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
No
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: biomarkers in oncology, including colorectal cancer
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |
|---|---|
| 1 | |
|
Version 1 21 Dec 21 |
read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)