Keywords
Deep Learning, Segmentation, Image Analysis, Interactive Machine Learning, ImJoy
This article is included in the NEUBIAS - the Bioimage Analysts Network gateway.
This article is included in the Bioinformatics gateway.
This article is included in the Software and Hardware Engineering gateway.
This article is included in the Artificial Intelligence and Machine Learning gateway.
Deep Learning, Segmentation, Image Analysis, Interactive Machine Learning, ImJoy
Deep learning-based methods have been widely used to analyze biomedical images for common tasks such as segmentation1,2, denoising3,4 and classification5. Despite their potential, building user-friendly deep learning tools and distributing them to non-experts remains challenging. Some attempts to tackle this challenge, like ImageJ plugins3,6,7 or web applications8–10, have been proposed. Among them, due to practical and technical challenges, most tools (e.g. ImageJ plugins) only allow for inference with pre-trained models. While it lowers the barriers for users to test and evaluate advanced tools, pre-trained models can easily fail or generate unexpected artifacts due to mismatch of the data distribution for many applications. Depending on the training data distribution and many other factors, applying pre-trained deep learning models can easily suffer from overfitting or other generalization problems. This makes them vulnerable to subtle changes such as noise patterns generated by different microscopes, or morphological changes from different samples. Therefore, it is important to improve the generalization of the pre-trained models, or to re-train and finetune models with users’ own data. Previous work including nucleAIzer10 and CellPose1 for nuclei and cell segmentation have demonstrated that more generalized models can be trained by using a large amount of labelled data covering many possible variations such as different imaging modalities and object types. The resulting pre-trained models show promising generalization capability and robustness when applied on unseen data without retraining. However, the fact that this requires a much larger and richer training dataset prevents its use in cases where such a dataset is too expensive to obtain. For most cases, it is thus required to re-train or fine-tune models with a user’s own data.
In a typical workflow for training a deep learning model, the first step is to annotate the objects (e.g. cells) in the image manually and obtain the corresponding masks. Typically, this step is labor intensive and time consuming. The second step is to use the labelled images to train a deep learning model (e.g. a U-Net). After training, one can use the trained model to process new images. Despite some metrics such as the Jaccard similarity coefficient which can be used to monitor the training, a common issue is that it is hard to predict how much annotation is required for training the model. As a result, users often need to iterate several rounds between annotation and training.
Meanwhile, interactive machine learning tools such as ilastik11 can be used to interactively annotate data and train models by combining the two steps in a more efficient way. It works by first annotating a small amount of data to allow the model to start training in the background. While the model is training, users can work on the annotation and add newly annotated data to the training dataset. In a later stage, instead of annotating all the objects in the image, users can use the model to obtain predicted objects and decide to accept or re-annotate them. This greatly improves the efficiency of both annotations and model training, which not only reduces the overall amount of work required but makes the new annotations more targeted to the weaker part of the model. By coupling the processing of active learning12, the total amount of training data can be further reduced. Besides that, the whole process is beneficial for users in helping them to understand the failure modes of the model.
Despite these advantages, many existing interactive machine learning tools are built for classic machine learning methods such as random forest. While ongoing efforts have been made to train deep learning models interactively, building tools that are accessible for non-experts remains challenging due to the increased complexity for annotating data and training deep learning models from an interactive graphical interface. Nevertheless, there have been many existing tools such as Jupyter notebook and many libraries in the Web and Python ecosystem that can be used for building interactive training processes. To combine them and make it even easier to work with, we developed ImJoy8 which is a web application for building interactive and scalable data analysis tools (ImJoy, RRID: SCR_020935).
In this work, we demonstrate a tool we built with ImJoy for interactive deep learning-based image segmentation. From a web based graphical user interface (GUI), it allows interactive annotations of images and training powerful deep learning models including basic U-Net and CellPose. While the GUI can run in a desktop web browser, a mobile phone, or a tablet with touch screen, the computation server part can run in a local workstation or remote server. In addition, the GUI can be embedded in Jupyter or Google Colab notebooks to allow easy customization and further development for the developers. It also works independently as an ImJoy plugin which can be easily shared with a hyperlink to the end users.
The tool is implemented as two ImJoy plugins13: a model trainer plugin and an image viewer plugin. The trainer plugin is mainly responsible for performing model training and the viewer plugin is built on top of Kaibu and used for visualizing and annotating the images. There are additional panels in the GUI for controlling and monitoring the trainer. The model trainer plugin requires Python (version 3.7+) along with other python modules, the image viewer plugin runs in a web browser which requires Chrome (version 80+) or Firefox (version 73+). The two plugins can run directly in ImJoy using Jupyter notebooks with ImJoy Jupyter extension installed, or in a Google Colab notebook.
As shown in Figure 1, the image viewer interface consists of 1) a set of tools for drawing or editing polygons to mark the objects in the image; 2) the image along with the annotation markups shown as different layers; and 3) control panels for listing the samples, controlling the trainer and monitoring the training loss. The viewer can run independently in modern web browsers on a desktop computer or tablets. Meanwhile, the trainer plugin is written in Python and can run through any Jupyter notebook server locally or remotely. Depending on the models, we use either Tensorflow or Pytorch as the base framework. If available, graphical processing units (GPUs) can be used for acceleration. The two plugins are connected via the remote procedure calls (RPC) provided by ImJoy such that the plugins can call each other’s functions and pass data between them in a transparent manner. For example, the viewer plugin can call the predict function in the trainer plugin to infer the labels and the trainer plugin can call a function in the viewer to obtain the user corrected labels. For interactive image annotation, a powerful combination is to use a touchscreen device (e.g. tablet or mobile phone) with a pen to draw the markups, and in the meantime, train the model with a remote Jupyter notebook server with GPU.
In addition to the ImJoy interface, with our newly developed Jupyter notebook and Google Colab extension for ImJoy, this interactive annotation tool can also be embedded directly in a notebook interface. This allows developers to quickly customize the trainer plugin in a notebook environment, debug and interact with the trainer through Python code. By running the tool on Binder14 or Google Colab with free computing resources (including GPU access), it allows users and developers to share and reproduce interactive workflows without setting up a local computational environment.
In order to use the tool, the user needs to prepare a set of unlabeled images and organize them into folders as required by the trainer. Then the user should open the plugin with ImJoy (version 0.11.29+) in a web browser such as Chrome (version 80+) and Firefox (version 73+) to see the image annotation interface. To run the trainer plugin, the user needs to connect to a local or remote Jupyter notebook server (Miniconda 3 or Anaconda 3 is recommended for installing the Jupyter notebook server). GPU hardware is recommended but not mandatory. Alternatively, the user can also use the annotation tool in a Google Colab notebook with GPU runtime type selected, and this is recommended for evaluation without any local installation.
To train a model from scratch, the user can start by loading an unlabeled image into the viewer and start the initial annotation process (Figure 2a). With the markup tool, the user can then draw polygons to outline each object (e.g. cell) in the image. Once done, these polygons will be sent to the trainer plugin and be saved into a text-based format named GeoJSON. Importantly, the image along with the GeoJSON annotation will be moved and added into a training sample pool. After annotating a few images, the user can start training the model. In another thread, a training loop will be started and for each iteration, the trainer will randomly take a batch of samples (e.g. 1–3 images) from the training sample pool, and train the model for one iteration and repeat the process until the user stops the training loop. After being trained for some iterations (e.g. in 2–5 minutes with a GPU from scratch), the user can start to use the model to perform predictions. From then on, instead of annotating the entire image, the user can send the image to the trainer to obtain the predicted labels, fix the wrong labels and keep the correct ones. Since the label correction process is typically much faster than annotating from scratch, the entire workflow is accelerated. The curated labels will be added to the training sample pool and used for training. Video 115 is a screen recording for using our tool to annotate images and train a CellPose segmentation model from scratch in Google Colab.
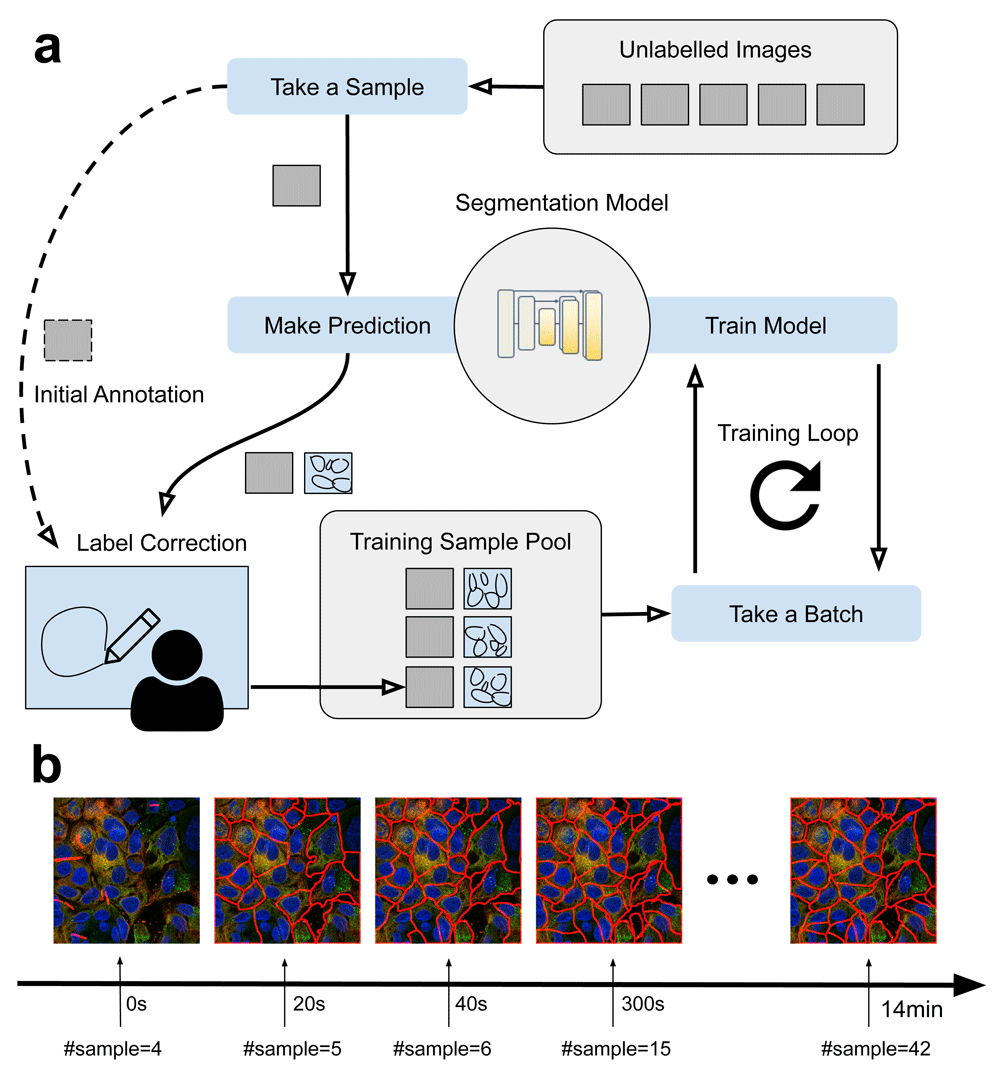
(a) Interactive annotation and training workflow. Starting with a set of unlabelled images, annotations are gradually made and moved to the training sample pool for training the model. After some iterations, instead of annotating images from scratch, the model can be used to make predictions such that the user can quickly correct the labels. (b) Segmentation results showing the progression of the interactive training. Specifically, a CellPose model being trained from scratch with 4 images at time 0, then we gradually add new samples to the training sample pool (20s/image). We test the model with a fixed image which is not seen by the network.
The trainer supports different types of segmentation models including U-Nets with various types of encoder variations16 and the CellPose1 model. It is worth noting that when a suitable pretrained model is used as a start, the user can also skip the initial annotation process and start to correct predicted labels.
The tool can be generally applied for annotating images and training deep learning models for segmentation. It is easy to customize and extend to support different types of data organization, image formats and model architectures.
For demonstration purposes, we provide a small example dataset17 from the Human Protein Atlas18 with 163 samples in total. Each sample is a 4-channel (microtubules, endoplasmic reticulum (ER), nuclei and protein of interest) cell image with manually annotated cell mask in GeoJSON format. The samples cover ~20 different cell-line types with variations in morphology.
For the model training part, we used a slightly modified CellPose model and training process. Specifically, we switched off the style connection in the U-Net used by CellPose and used Adam19 as the optimizer. The same rotation and scaling augmentations were used as in the original CellPose training workflow. Only two channels (ER and Nuclei) were used as input to the model. Starting by using four labelled images in the training sample pool, new images and labels were gradually moved to the training sample pool during training. Figure 2b illustrates the rapid progression of the interactive learning process. Specifically, we started the trainer (at time 0) and gradually added one annotated sample every 20s into the training pool. With the same test image, we checked the predicted labels. As shown in the figure, the result rapidly improved in less than a minute. In order to better understand the learning process, we initialized the model with random weights (i.e. no pretrained model is loaded). Compared to a conventional annotation workflow, the interactive tool can accelerate the annotation by roughly 6 times (it takes ~2 minutes to manually annotate the image from scratch, and ~20s to correct labels with our tool).
We demonstrated an interactive annotation and training tool that is capable of accelerating the annotation process for image segmentation. It works by running a training loop in the background while adding new annotations. The feedback loop improves the annotation efficiency and allows more flexibility for the user to control the training and decide on whether new annotation is needed. With the example dataset, we demonstrated improved efficiency for training and annotation. This type of incremental process is more user-friendly for people who want to adopt deep learning-based methods.
We foresee that the developed model can be further improved to increase its efficiency, reproducibility and areas of applicability. A more advanced version could for example support 3D annotation and segmentation, allow recording of the annotation and training activities, or use more advanced sampling schemes when selecting which images the user should annotate (also known as active learning).
Zenodo: HPA Cell Image Segmentation Dataset. https://doi.org/10.5281/zenodo.443089217
This project contains the following underlying data:
Sample dataset, source codes and tutorial are available from: https://github.com/imjoy-team/imjoy-interactive-segmentation
Archived source code at time of publication: https://doi.org/10.5281/zenodo.446108013
Data are available under the terms of the Creative Commons Attribution 4.0 International license (CC-BY 4.0).
We thank Christian Gnann for testing and evaluating the performance of the interactive annotation tool. The example dataset is annotated by Jay Kaimal and Peter Thul.
This publication was supported by COST Action NEUBIAS (CA15124), funded by COST (European Cooperation in Science and Technology).
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the rationale for developing the new software tool clearly explained?
Yes
Is the description of the software tool technically sound?
Partly
Are sufficient details of the code, methods and analysis (if applicable) provided to allow replication of the software development and its use by others?
Partly
Is sufficient information provided to allow interpretation of the expected output datasets and any results generated using the tool?
No
Are the conclusions about the tool and its performance adequately supported by the findings presented in the article?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Machine learning, computer vision, bioimage analysis, computational neuroscience
Is the rationale for developing the new software tool clearly explained?
Yes
Is the description of the software tool technically sound?
Yes
Are sufficient details of the code, methods and analysis (if applicable) provided to allow replication of the software development and its use by others?
Yes
Is sufficient information provided to allow interpretation of the expected output datasets and any results generated using the tool?
Yes
Are the conclusions about the tool and its performance adequately supported by the findings presented in the article?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: image analysis, microscopy, imaging
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 24 Feb 21 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)