Keywords
Cell Fate Engineering, Chemical reprogramming, Direct cell conversion, Transdifferentiation
This article is included in the Bioinformatics gateway.
This article is included in the Cheminformatics gateway.
Cell Fate Engineering, Chemical reprogramming, Direct cell conversion, Transdifferentiation
Cell engineering technologies possess a tremendous potential for basic and applied science. Recent discoveries in this field pushed forward regenerative and personalized medicine, drug discovery, and toxicology. In the future, cell engineering technologies can enable regeneration of altered tissues and extend the human lifespan. Now they serve as a platform for drug testing and toxicology experiments. Furthermore, these approaches enable molecular mechanisms to be revealed that drive normal development and differentiation. Transcription factors (TFs) have been used to manipulate cell fate in the majority of published protocols.1 Low efficiency and safety concerns restrict the application of cell fate engineering approaches in clinical practice.2 Small molecules that target signaling pathways or regulate the epigenetic state offer powerful tools for refined manipulation of cell fate to the desired outcome.3 There is an increasing number of protocols that utilize small molecules in order to induce lineage differentiation or to facilitate transdifferentiation by increasing efficiency or by replacing genetic reprogramming factors.
Small-molecule-mediated approaches have more potential for clinical applications since:3
1. The biological effects of small molecules are typically rapid, reversible, dose-dependent and predictable, allowing precise control over specific outcomes and precise resembling of target cell type;
2. Compared with genetic interventions, the relative ease of handling and administration of small molecules make them more practical for further therapeutic development;
3. Small molecules do not evoke safety concerns about possible genome integration in contradistinction to genetic interventions.
Chemical cell conversion protocols vary not only in chemicals used for reprogramming but also in media, making it difficult to choose the optimal protocol in practice. A database integrating cellular reprogramming protocol has been already published.4 Yet, the protocol representation is not standardized and the database has not been updated recently. Here emerges the need for aggregation of protocols in a standardized form that would contain all relevant and specific information from them. Also, aggregators should provide user-friendly access to enable expeditious retrieval of information.
In this work, we present a protocol aggregator, CFM (cell fate mastering), with a user-friendly interface that allows stem cell biologists to compare protocols, select the best one for their needs with high efficiency and reliability as well as leave feedback for their experience regarding the protocol for the research community. Additionally, we provide information about a potential mechanism of drug action and pathways involved in cell conversion. Data on molecular mechanisms affected by specific compounds could help to improve the protocol. We use expert opinions to develop an original standard for protocol representation, provide public ratings for protocols calculated based on users’ evaluation, and maintain a discussion forum. Protocol ratings as well as detailed feedback from the community will help to promote high-quality and reproducible protocols. Standardized and detailed protocol representation will facilitate development and validation of systematic computational approaches for cell fate engineering (e.g. DECODE5). The current version of CFM contains 169 distinct protocols, 113 types of cell conversions, and 158 small molecules. Summary statistics are available in Figure 1.
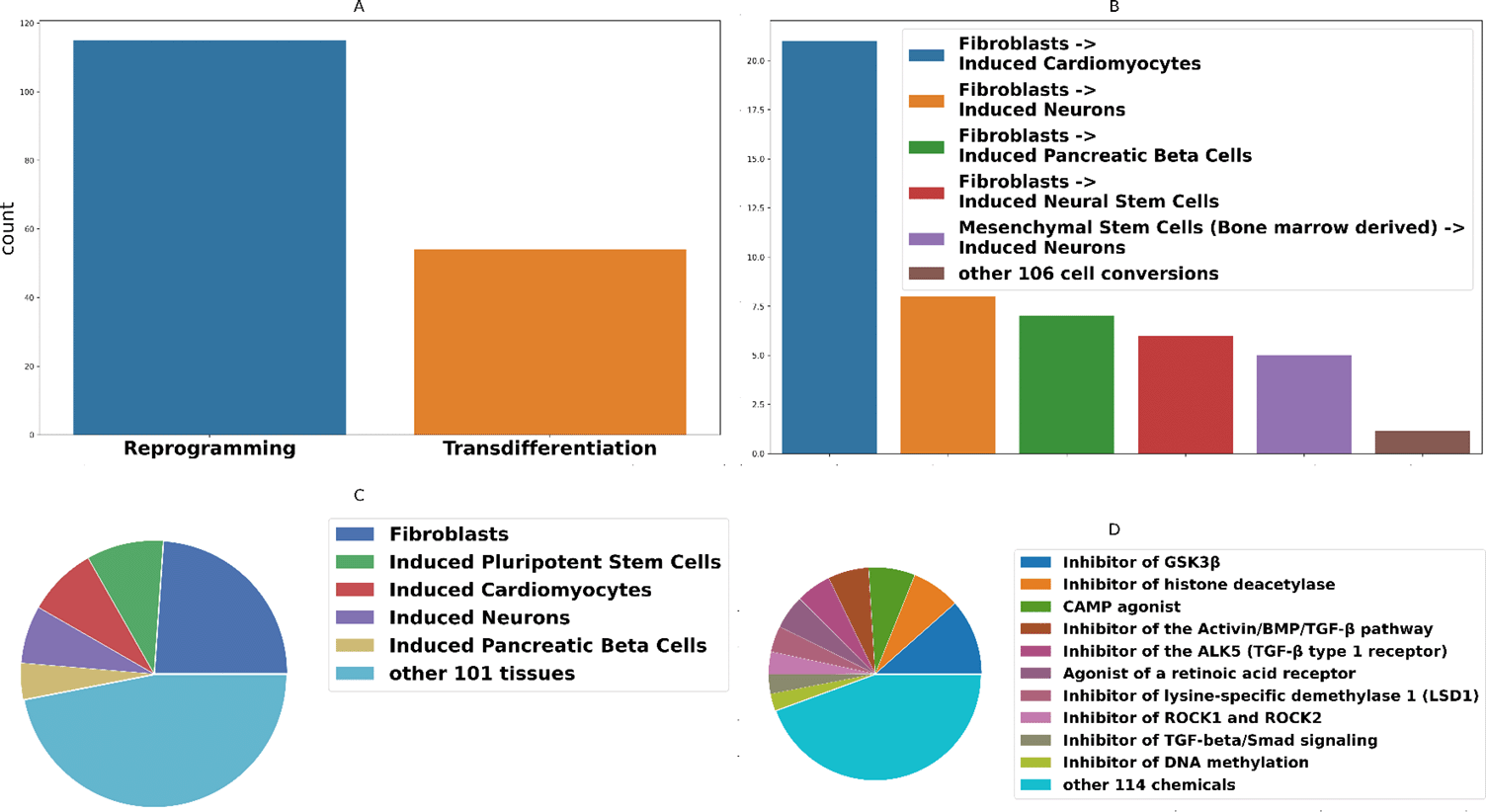
(A) Number of types of cell conversions. (B) Number of cell type conversion pairs in CFM (the bar for ”other 106 cell conversions” represents an averaged number of a protocols for each other cell conversions type). (C) Frequencies of chemical compounds used in the protocols. (D) Frequencies of initial cell types used in protocols.
We are committed to a strong database maintenance strategy, which means:
1. We manually check data in order to provide comprehensive information. Public rating is conducive to promoting reproducible protocols;
2. We communicate with experimental biologists and permanently improve and extend the functionality of the database to meet their needs. Hence, we created a special feedback form where users can inform us what features they would like to add;
3. We support protocol updating by users (after manual curation of the suggested protocol);
4. We also plan to update the content of the database on a regular basis (at least bi-annually).
We collected protocols from PubMed and Google Scholar using the keywords ‘direct reprogramming’ AND ‘chemicals OR small molecules’, ‘transdifferentiation’ AND ‘chemicals OR small molecules’, ’direct cell conversion’ AND ‘chemicals OR small molecules’. In this way, more than 1000 papers were obtained. Experts prefiltered papers based on the content of key sections (abstract, introduction, method) discarding irrelevant papers. We manually extracted information from each relevant article and added data about small molecules implicated in protocol from PubChem as a part of postprocessing. The overview of the whole workflow can be seen in Figure 2.
The database can be queried at cfm.mipt.ru/query page (Figure 3). By default, all protocols are listed. If no protocols match a query, the whole database is displayed (default view). Search is case-independent. The first column contains a link to protocol description in the cfm.mipt.ru/viewProtocol/<protocolId> format, where protocol ID is an inner identifier. The protocol description page contains article information (DOI, article title, and authors), source and target cell lines , initial cell culture description, the species from which the source cell line was obtained, the total protocol duration, the media in which the protocol was implemented, protocol yield (with comments on how it was calculated), chemicals (with their methods of effect is known), transcription factors, stress factors and growth factors used during the procedure and some comments on the protocol in general. Also, we provide a simple interface for rating protocols for registered participants. After curation, the rating will be available on the protocol page. All rated protocols are shown in a personal user account (cfm.mipt.ru/login). A researcher can add a protocol (and a published related paper) to the CFM database by submitting it through cfm.mipt.ru/add after the email is verified by a CFM administrator. Detailed information about verification can be found on the Add Protocol page.
We strongly believe that the development and maintenance of such a database will encourage researchers to keep improving protocols for cell conversion. Among all cell conversions, reprogramming protocol still dominates over trans-differentiation Figure 1. As it is clear from the Figure 1B and C, fibroblasts are widely used as an initial cell type in various protocols, while other cell types are underrepresented. Although fibroblasts might be an easy cell type to obtain in the clinic, other initial cell types such as blood cells could be also of use. As can be seen in Figure 1D, there is a small group of chemicals effective in several protocols, suggesting that they target a general mechanism. Possibly other chemicals from the same functional class, such as epigenetic drugs, should also be tested for their cell conversion potential. Further research on the regarding relations between cell conversions and chemicals, media, transcription factors, etc. should lead to the development of a computational framework that will facilitate acceleration of the search for conditions conducive to new cell conversions which are highly applicable in medicine (for example,6-8).
In Figure 4, as well as on Figure 3, you can see examples of querying the database. New fields of search can be added using the green ADD button; unnecessary fields can be removed using the red DELETE button.
We calculate the similarity of molecules based on their SMILES. Retrieving Morgan fingerprints from them, we calculate the Tanimoto similarity and provide the result to the user. The interface can be seen in Figure 5.
All data was gathered from open sources such as PubMed; links to the source articles can be found on protocol pages accessible from cfm.mipt.ru/query page.
We collected protocols from PubMed and Google Scholar, and downloaded SMILES for chemical compounds from PubChem. All data can be downloaded from our GitHub Repo.
Our service is available at: http:cfm.mipt.ru
Source code is avaliable from: https://github.com/lubitelpospat/CFM-source
Archived source code as at time of publication: http://doi.org/10.5281/zenodo.4500125.9
License: MIT
The authors are grateful to Valeria Tsvelaya for her help with the standardized description of the protocols. We are also grateful to the creators and maintainers of https://pubchem.ncbi.nlm.nih.gov.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the rationale for creating the dataset(s) clearly described?
Yes
Are the protocols appropriate and is the work technically sound?
Yes
Are sufficient details of methods and materials provided to allow replication by others?
Yes
Are the datasets clearly presented in a useable and accessible format?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Cell reprogramming, tissue engineering
Is the rationale for creating the dataset(s) clearly described?
Yes
Are the protocols appropriate and is the work technically sound?
Yes
Are sufficient details of methods and materials provided to allow replication by others?
Yes
Are the datasets clearly presented in a useable and accessible format?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: cell biology
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 16 Apr 21 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)