Keywords
mass spectra, astrocytoma and glioblastoma tumors, dimensionality reduction, classification, high- and low-resolution
This article is included in the Artificial Intelligence and Machine Learning gateway.
This article is included in the Oncology gateway.
mass spectra, astrocytoma and glioblastoma tumors, dimensionality reduction, classification, high- and low-resolution
The extent of tumor resection is important for patients with primary brain tumors in terms of life expectancy since tumor cells can provoke a disease recurrence1. Recently, we have seen a growing interest in the use of mass spectrometry for the identification of tumor tissues, typing, and detection of tumor boundaries2-4. Analysis of tumor samples using mass spectrometry is based on the observation that tumor cells differ significantly from normal ones in their metabolic processes and, as a consequence, have a different chemical composition5-8. Identification of the histological type and location of the brain tumor tissue during neurosurgical intervention allows for the correct dissection of the tumor and opens the way to a personalized strategy for further treatment of the patient with chemotherapy, taking into account the molecular features of the tumor. Comparative analysis of tumor types is fundamental, although elucidating tumor boundaries is of the highest priority for neurosurgeons9.
Fast mass spectrometric profiling allows rapid clinic and laboratory analyses but faces the problem of classification of high-dimensionality objects.
For the analysis of large mass spectrometry (MS) data, dimension reduction (DR) algorithms are commonly used as a previous step for statistical analysis and visualization. The most widely used DR methods are linear methods such as PLS-DA and PCA10-16. More advanced nonlinear methods have recently been developed, such as t-SNE and UMAP, to name a few17. DR methods allow visualization through the major components of the compressed data, e.g. the first three PCA components and three selected ions are the most commonly used characteristics in MS imaging18,19.
Despite the visualization that could produce the results dividing the different types of tissues by their MS, these results are subjective and require further automated classification. Machine learning (ML) methods are commonly used for these purposes20-22.
In this paper, we compare the performance of six DR algorithms and ten ML algorithms in their ability to classify the MS profiles of astrocytoma and glioblastoma. Also, we compared the stability of the trained ML models on the data obtained with another instrument and under different conditions. The stability of the trained models to different instruments is very important for the wide spreading of the methods, as the clinical conditions could influence the mass spectra due to different mass analyzers. Different polarities, resolutions, and m/z ranges are also considered.
The two instruments used in our study: for both high and low resolution (HR and LR) under laboratory conditions (Thermo LTQ Orbitrap XL) and only low resolution under clinical conditions (Thermo LTQ XL). Inline cartridge extraction23 followed by electrospray ionization was used for mass spectrometric profiling of samples. Spectra from Thermo LTQ Orbitrap XL were measured in both positive and negative ion modes with two resolutions: 24,000 at m/z=744 (high resolution using Orbitrap analyzer) and 2000 at m/z=744 (low resolution using LTQ analyzer). All spectra were measured in the m/z 500 — 1000 and m/z 100 — 2000 ranges.
Tissue samples were provided by the N.N. Burdenko NSPCN and analyzed under a protocol approved by N.N. Burdenko NSPCN Institutional Review Board (order 40 from 12.04.2016, revised with order 131 from 17.07.2018). A signed informed consent form, filled out in accordance with the requirements of the local ethical committee, specifically noting that all removed tissues can be used for further research, was obtained from all patients before surgery. The study was conducted in accordance with the Helsinki Declaration as revised in 2013. All procedures were carried out according to the relevant guidelines and regulations.
Three fragments of tissue taken from a single patient were measured with each instrument to take into account and evaluate inner biological variability. In total, 76 astrocytoma fragments (26 patients) and 89 glioblastomas (31 patients) were measured with Thermo LTQ XL (Ltq) in LR, while 60 astrocytoma fragments (20 patients), 84 glioblastoma fragments (28 patients) were measured with Thermo LTQ Orbitrap XL (Orbitrap) in LR and HR. Samples from 37 patients were measured with both instruments, 20 patients were measured only with Ltq, 11 — only with Orbitrap. The precise schema is given in Figure 1.
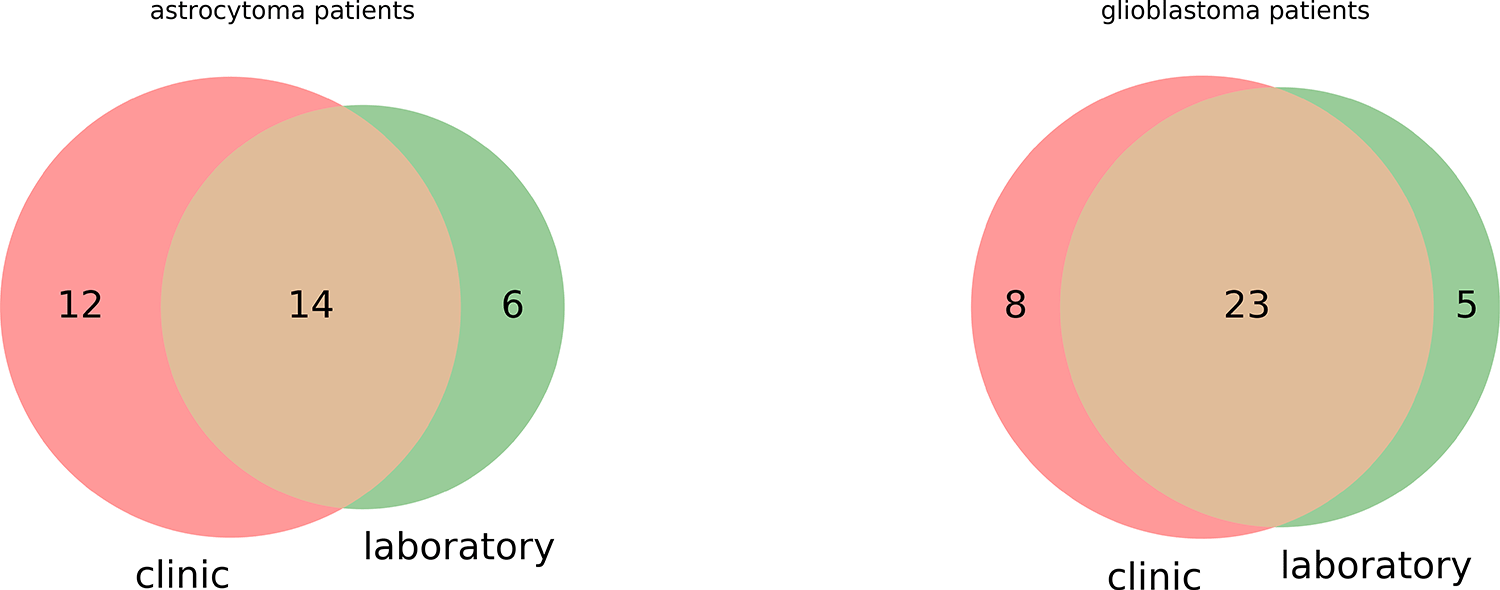
There tissue samples (total 309) from 68 patients had different brain tumors: 21 anaplastic astrocytomas (WHO Grade III; 9 tumors with IDH-1 R132H mutation), 10 diffuse astrocytomas (WHO Grade II; 7 tumors with IDH-1 R132H mutation), 1 gemistocytic astrocytoma WHO Grade II (IDH-1 with IDH-1 R132H mutation) and 36 glioblastomas (WHO Grade IV; 11 tumors with IDH-1 R132H mutation).
Tissue samples were divided into several parts, for annotation and MS analysis. Tissue samples were annotated with routine hematoxylin and eosin staining and further immunohistochemical analysis of its fragment (expression of isocitrate dehydrogenase 1 (IDH-1)). Other fragments of tissue samples were measured with Thermo LTQ XL right after removal or were placed in the normal saline, frozen, and stored at -80°C until measurement with Thermo LTQ Orbitrap XL. Measurement for each sample was carried out continuously with alternating mode, range, and resolution if possible24.
Spectra were processed with the algorithm similar to described previously25,26. Mass spectra were binned with binning width 0.01 m/z, and then spectra were convoluted with Gaussian (FWHM equals 0.4m/z for high-resolution and 0.2m/z for low-resolution). Spectra of each measurement were filtered by a moving median filter. The width and step of the median filter were chosen to 51. The baseline subtraction was carried out through Kneen and Annegarn’s algorithm27. Spectra similarity matrix (SSM) with cosine measure similarity was calculated as described previously26. Both dimensionality reduction and classification were made with Scikit-learn v0.23.128 machine learning library.
All calculations and visualizations were made using code written by the authors using MATLAB R2019b (GNU Octave 5.2.0 could be used for reproducing results) or Python 3.7. This code is freely available at http://doi.org/10.5281/zenodo.430770043.
The classification without dimensionality reduction is compared with the method of principal component analysis (PCA), non-negative matrix factorization (NNMF)29, isometric mapping (Isomap)30, Partial least squares discriminant analysis (PLS-DA)31, UMAP32, Diffusion map33. The number of the selected features after dimensionality reduction equals 5.
Nearest Neighbors34, linear support vector machine SVM35, Radial-basis function (RBF) kernel SVM35, Gaussian Process36, Decision Tree37, Random Forest38, Neural Net (Multi-layer Perceptron classifier with 1 hidden layer with 100 neurons and the log-loss function and “adam” optimizer and L2 regularization), AdaBoost39, Naive Bayes40, QDA41 were used as classifiers. Only one scan from each measurement after median filtration was taken part in the classification task.
SSMs of astrocytoma and glioblastoma in different polarities are presented in Figure 2. There are two clusters of fresh and frozen samples. The first were measured right after surgery, the second after storing samples under −80°C. Figure 2 represents the large-range data (100-2000 m/z) measured in low-resolution.
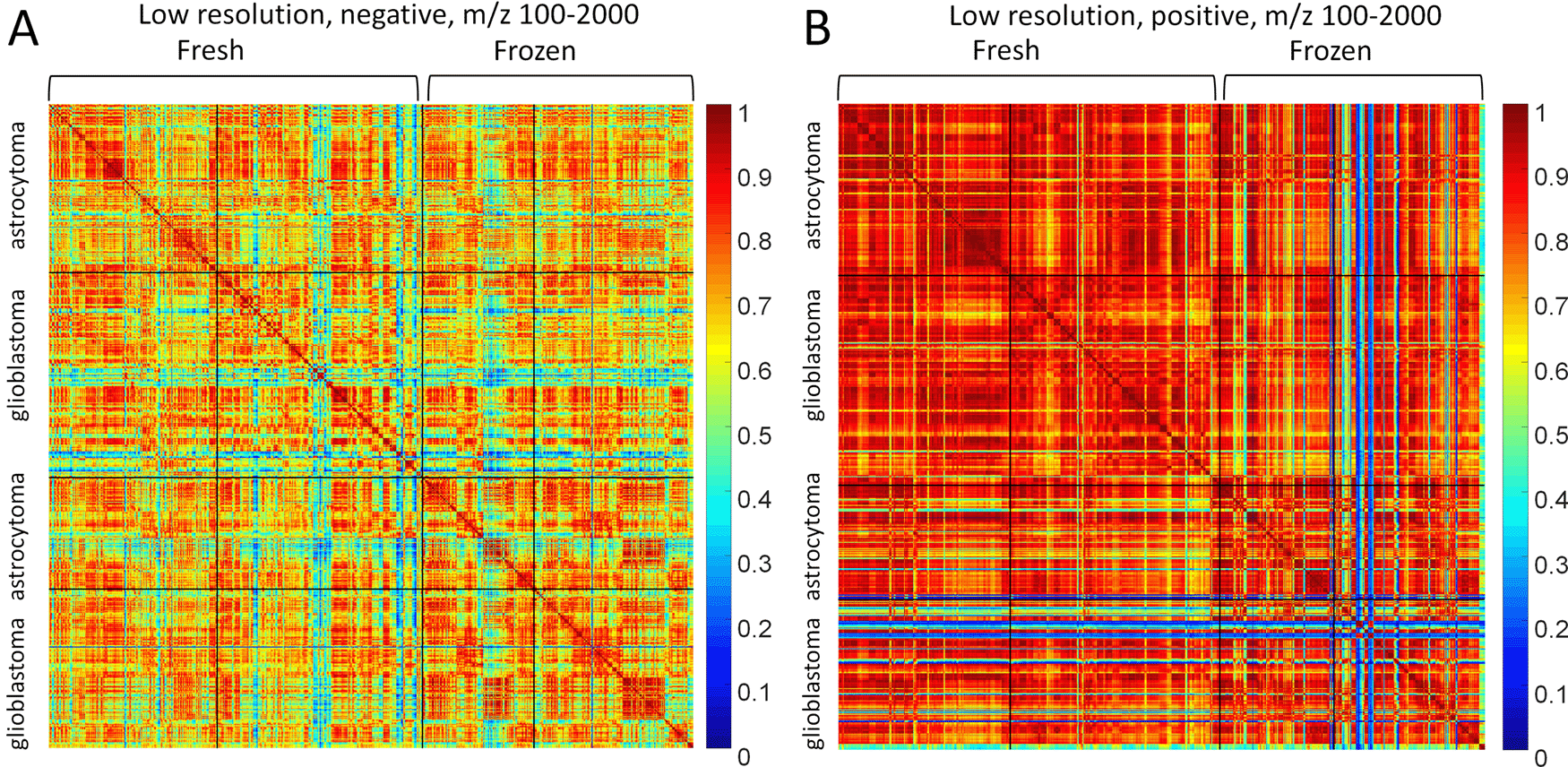
(A) Negative and (B) positive mode.
Frozen glioblastoma MS profiles have a lot of outliers, which is obvious from the SSM Figure 2B. The outliers could be filtered out from the datasets for the classification task, but the classifiers will be created with only fresh samples. Thus, outliers will be tested on the trained classifiers and the results of visual inspection of SSM could be confirmed. The negative mode (Figure 2A) has more variance and spectra of the negative mode have less similarity. At the same time, there are no obvious differences between astrocytoma and glioblastoma. We, therefore, employ the machine learning methods for classifying the MS profiles.
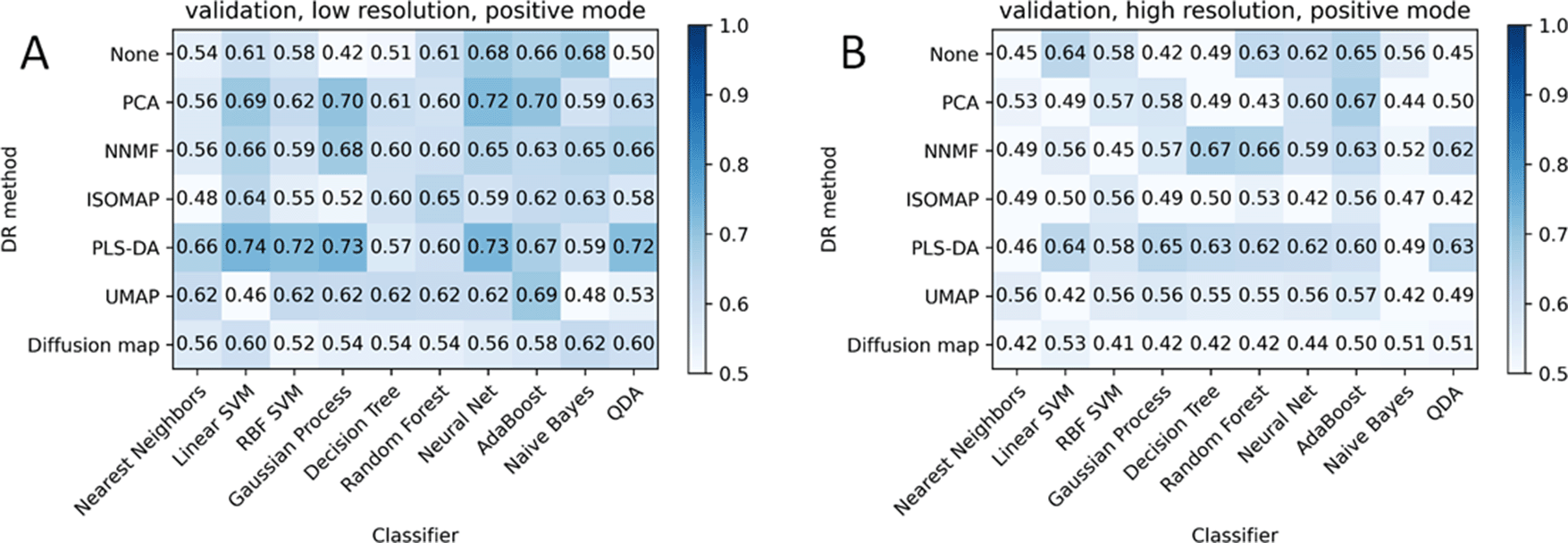
The accuracy of classifiers for different dimensionality reduction algorithms is presented in Figure 3. The accuracy was calculated as the ratio of correct predictions to the total number of predictions on the given data and labels.
First 70% of astrocytomas and 70% of glioblastomas were taken as the training data, the rest as the test data. Neither of the mass spectra of the samples corresponding to the single patient was in training and test data simultaneously to prevent the overfitting. Thus 46% of training data and 46% of test data were astrocytoma class, whereas 54% of the training and test data were glioblastoma class. Many of the combinations of classifiers and dimensionality reduction algorithms score about 46% or 54% of accuracy.
The PLS-DA is overfitted in all cases except probably negative mode and m/z 500-1000 (Figure 3E and F). The highest values of accuracy are achieved with NNMF for positive mode MS profiles. NNMF produced better results for a wide m/z range (100-2000) since classifiers produce similar accuracy in this case. The best result of NNMF and random forest combination for the positive mode, m/z 500-1000 is achieved for 5 components of NNMF. The most stable results were produced for the 100-2000 m/z range in positive mode. These conditions are considered below
The robustness of each combination of DR and ML algorithms is tested on the data obtained with another instrument (Orbitrap) both in low and high resolution (Figure 4). In this setup, the classifiers were taken from the previous step, so they were fitted with Ltq data. High-resolution data was roughed through Gaussian convolution to decrease resolving power to low-resolution data.
(A) Low-resolution; (B) high-resolution converted to low-resolution.
Figure 4 demonstrates the lack of classifier robustness to the different instruments despite the rather high similarity of MS profiles from Figure 2. The combinations of the algorithms that seem to be overfitted from Figure 3C and D (Diffusion map, PLS-DA) leads to almost random classification for both low-resolution and high-resolution converted to low-resolution data.
Considering data without outliers provides better results (Figure 5) for overfitted models, whereas it has an almost negligible influence on the other models’ accuracy. The exclusion of outliers with the SSM by indicating low-similarity blue strips mostly in glioblastoma samples reduces the number of samples from 144 to 116 for low-resolution data and from 144 to 113 for high-resolution. It increased the accuracy, for example, of NNMF+Linear SVM by five percent.
The combination of the PCA and AdaBoost approach demonstrates here the most robustness to the instrument.
The SSMs in Figure 2 demonstrate different variance of MS profiles in negative and positive mode. The negative mode has less similarity between MS profiles especially in the m/z range 100-2000. The positive mode has similar MS profiles excepting the outliers that are not contained in the negative mode24,42. Thus, the positive mode could be used for looking for general distinctions between classes, whereas the negative mode could contain fine distinctions in the class as well. The classifiers seem to have difficulties in application to such high variation objects as MS profiles have in negative mode.
Preliminary feature selection (dimensionality reduction) is necessary for classifiers to not be overfitted, as the dimensionality of the object is equal to 7600 features in the given preprocessing (binning for m/z 100-2000 range) and the number of samples is about 150 (about 50 patients) in the dataset from the single instrument. The dimensionality reduction is expected to work rather effectively in the first several components only in case the variation is caused by interclass variability with classes characterized by a single Gaussian distribution. In the presented case, each class has a set of multi-dimensional Gaussian distributions. Also, astrocytoma tissues can have smooth changes of grades up to glioblastoma due to its origin. The whole variability of the data could be explained by six factors: tumor type, tumor localization, mutation status, interpatient variability, intratumor variability, and batch effect. Usually, interpatient variability is comparable with interclass variability. So, the presented data is unlikely to have most of its variability in the first components due to tumor type. Thus, both for the data with low (positive mode, Figure 2), and with high variance data (negative mode Figure 2) the first components of DR could be useless in terms of classification if the intraclass variability is higher than interclass.
PCA (Figure 3) provides good results for positive mode: most classifiers achieve similar accuracy of about 80%. PCA is ineffective for negative mode, as the accuracy falls to ~46%/54%, which corresponds to the ratio of the classes in the dataset. At the same time accuracy in the training data is much higher, so PCA provides specific only for the training data, which leads to overfitting. PCA and further classifiers predict mostly one class for any test data in negative mode.
NNMF components correspond to the highest accuracy values in most conditions. It seems to be not overfitted in positive mode. NNMF produces the best results for the m/z range 500-1000 in combination with random forest or decision tree models. NNMF and PCA repeat the accuracy of the test data with the KNN classifier for positive mode.
UMAP seems to be perspective, whereas Isomap, Diffusion map, PLS-DA algorithms demonstrate worse results compared with others. Classifiers are the most overfitted if no DR algorithm is applied, as expected. Although the AdaBoost model has about 70% accuracy on the test data for each experimental condition for non-preprocessed data (Figure 3).
PLS-DA is used for dimensionality reduction here, but it is common practice to use it as a classifier. PLS-DA appears to be overfitted for all cases except the 500-1000 m/z range in negative mode, where all other combinations are overfitted. Random forest demonstrates the best result for this case but still seems to be slightly overfitted, as the accuracy on the test data decreases by 20% compared with accuracy on the train data.
Neural net and AdaBoost models demonstrated the best results for uncompressed data in negative mode.
Algorithms were applied to a similar dataset of frozen samples, measured with another instrument (Orbitrap). As the measurements were carried out in two resolutions, the accuracy matrices are calculated for both resolutions. Figure 4 shows a lower level of accuracy compared to Figure 3, e.g. for PCA+AdaBoost. This is explained by the outliers, which could be seen in Figure 2B for frozen glioblastomas as the blue crossing stripes correspond to MS profiles that don’t have similar MS profiles in the dataset. Excluding the outliers from the consideration reveals the accuracy improvements, but the accuracy is still not so high. The combination PCA+AdaBoost demonstrated the most robust result of classification over all the data in positive mode (Figure 3-5). Thus, the most independent combination of dimensionality reduction and classification algorithm to the instrument, resolution, and freeze-thaw process is supposed to be PCA+AdaBoost. This combination accuracy is not changing for positive mode from train to test data on any instrument and equals to about 70-80%.
1. Positive mode mass spectra provide better accuracy for astrocytoma and glioblastoma classification by mass spectrometric profiles of samples without sample preparation.
2. Astrocytoma and glioblastoma could be classified with 88% accuracy in low-resolution (NNMF+random forest) by positive-mode mass spectrometric profiles.
3. The PCA and AdaBoost combination appeared to be most stable in positive mode while transferring the classifier from the Ltq’s to Orbitrap’s data. The accuracy of classification is about 65-70% for validation data.
4. The dimensionality reduction algorithms combined with the classification models can process the outliers from the SSM as normal data.
Zenodo: Data and code for comparison of different machine learning methods and dimensionality reduction for classification astrocytoma and glioblastoma tissues by mass spectra, https://doi.org/10.5281/zenodo.430770043.
This project contains the following underlying data:
Datasets of mass spectrometric profiles for different instruments, ranges, polarities, and resolutions.
Software files for figure replication.
Data are available under the terms of the Creative Commons Attribution 4.0 International license (CC-BY 4.0).
The research used the equipment of Shared Research Facilities of N.N. Semenov Federal Research Center for Chemical Physics of the Russian Academy of Sciences.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
Partly
If applicable, is the statistical analysis and its interpretation appropriate?
Partly
Are all the source data underlying the results available to ensure full reproducibility?
Partly
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: mass spectrometry
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
Partly
If applicable, is the statistical analysis and its interpretation appropriate?
Partly
Are all the source data underlying the results available to ensure full reproducibility?
Partly
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: mass spectrometry, proteomics, proteogenomics, molecular medicine
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 21 Jan 21 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)