Keywords
VEGF polymorphisms, allograft, renal, kidney transplantation, meta-analysis
VEGF polymorphisms, allograft, renal, kidney transplantation, meta-analysis
A, adenine; AR, acute rejection; C, cytosine; CA or C/A, cytosine/adenine; CEU, European population; CI, confidence interval; CR, chronic rejection; CRAD, chronic renal allograft dysfunction; C/T, cytosine/thymine; du, duplicate; G, guanine; GIH, Gujarati Indian population; GG or G/G, guanine/guanine; het, heterozygous genotype; HWC, Hardy-Weinberg Compliant HWE, Hardy-Weinberg Equilibrium; I2, measure of variability; ITU, Telugu Indian population; KT, kidney transplantation; LD, linkage disequilibrium; n, number of studies; NRJ, non-rejection; OR, odds ratio; Pa, P-value for association; Phet, P-value for heterogeneity; [R], reference of studies; RJ, rejection; SNP, single nucleotide polymorphism; T, thymine; var, variant allele or genotype; VEGF, vascular endothelial growth factor gene; VEGF, vascular endothelial growth factor protein; wt, wild-type allele or genotype
Chronic kidney disease is a longstanding global health problem with substantial effects on morbidity and mortality1. Even with medical intervention, the likely endpoints in the progression of this disease are end-stage renal disease and kidney failure. In such cases, kidney transplantation (KT) is the current best available therapeutic option1,2. Success of the transplanted organ or an allograft in the recipient is limited by graft rejection3 which is characterized by inflammatory responses toward the graft tissue resulting in structural and functional impairments leading to allograft dysfunction4. Allograft rejection can be categorized largely into acute rejection (AR) which occurs days/weeks up until three months post-KT, or chronic rejection (CR) which is seen as progressive loss of graft function after three months post-KT5. Key factors that contribute to allograft rejection may involve cytokines that are secreted by immune cells and antibodies against graft antigens6. Cytokines have been recognized as potent immunomodulatory biomolecules that mediate physiological and pathological immune responses. These molecules determine the magnitude of alloimmune responses after transplantation, which influence graft survival7. Differences in genetic background of transplant recipients are, in part, the cause of varying immune responses towards grafts8. Recognizing these genetic differences and their effects on the immune response may help establish individualized immunosuppressive regimens that can improve allograft outcome9. This is accomplished by identifying the alleles that may increase risk or confer protection for immune-mediated complications after KT10. Single nucleotide polymorphisms (SNPs) in the cytokine genes may impact graft survival by altering transcriptional activities and levels of gene expression11 which lead to variations in cytokine production12.
Of the cytokine factors related to immune-mediated renal graft injury, the vascular endothelial growth factor (VEGF) is of potential use as a post-transplantation biomarker13. As mediator of vascular formation, VEGF promotes endothelial cell proliferation, differentiation and survival14. It also mediates endothelium-dependent vasodilation and maintains vascular permeability15. Dysregulations of VEGF expression are evident in many renal abnormalities16,17. This suggests a possible pathologic role of this protein in renal diseases including graft injury. Studies of allograft tissues from rat KT models (in both AR and CR events) and human KT recipients with AR showed increased VEGF expression in renal tubules and interstitium18,19. This suggests involvement of this gene/protein in the pathogenesis of allograft rejection. Various SNPs in the VEGF gene have been identified20,21 and reported to be associated either with low or high VEGF protein production21,22. One of the common VEGF SNPs, a cytosine (C) to adenine (A) polymorphism at position 2578 within the promoter region (-2578 C/A), was found to be associated with VEGF expression and allograft rejection. The CC genotype was associated with high VEGF production but varied in its effects on renal allograft outcomes with reduced23 and increased24 rejection risks across the studies. Given the varied influence of these SNPs on renal allograft function, it is opportune to statistically synthesize these study findings using meta-analysis.
Our study aims to provide better understanding of the genetic role of VEGF SNPs on post-KT allograft outcome in term of risk for allograft rejection among recipients, which might guide potential future directions in transplant genetics. To obtain less ambiguous, clearer estimates of the VEGF role in this investigation, we apply meta-analysis techniques (i.e. outlier treatment) in order to strengthen the evidence.
We searched for association studies on 13 February 2020, the start date for this meta-analysis. Four strings of search terms were used that included combinations of “vascular endothelial growth factor”, “VEGF”, “polymorphism”, “cytokine”, “renal”, “transplant”, “allograft”, and “kidney transplantation” as medical subject heading and text in MEDLINE using PubMed, Google Scholar, Science Direct and Mednar, unrestricted by language. Details of the search strategies for each of these four databases are shown in Table S1 (Extended data25).
References cited in the retrieved articles were also hand-screened to identify additional eligible studies. In case of duplicate articles, we selected the one with a later date of publication.
The following PICO elements were applied in the meta-analysis: (i) Population: renal allograft patients; (ii) Intervention: VEGF gene polymorphisms; (iii) Comparators: rejectors (RJ) versus non-rejectors (NRJ); and (iv) Outcome: allograft rejection post-KT.
Inclusion criteria were: (i) case–control design evaluating the association between VEGF SNPs and risk of allograft rejection; (ii) available VEGF genotype frequencies in the presence and absence of allograft rejection and (iii) sufficient genotype frequency data to enable calculation of the odds ratios (ORs) and 95% confidence intervals (CIs). Exclusion criteria were studies that: (i) did not involve renal allografts; (ii) were review articles; (iii) were functional studies; (iv) did not involve VEGF SNPs and with genotype or allele frequencies that were unusable/absent or, when available, combined with SNPs in other genes, preventing proper data extraction.
We examined four SNPs (Table 1; Extended data: S2 Table25). Observed phenotypic associations have been attributed to the proximity of SNPs in the VEGF gene26–28, termed linkage disequilibrium (LD). LD is the correlation between alleles located near each other29 and is measured in terms of D′ and r2 with a value of 1 indicating complete LD30,31. LD values were based on the European (CEU), and the Indian populations (Gujarati: GIH and Telugu: ITU) from LDlink. Complete LD between rs699947 (-2578C/A) and rs144854329 (-2549 insertion/deletion) merited combination, labeled VEGF1. -1154G/A (rs1570360), and 938C/T (rs3025039) were not in complete LD, thus analyzed separately, notated as VEGF2 and VEGF3, respectively (Table 2).
| First author | [R] | Year | Country | Ethnicity | Age (y) mean ± SD | Comparisons (/: versus) | VEGF polymorphisms (KT outcome) n | Clark- Baudouin score |
|---|---|---|---|---|---|---|---|---|
| Mittal | 39 | 2011 | India | Indian | 36.1 ± 10.2 | RJ / NRJ | rs699947, rs1570360 (AR) 2 | 10 |
| Prakash | 40 | 2015 | India | Indian | 37.1 ± 9.4 | AR / NRJ | rs699947, rs1570360, rs3025039, rs144854329 (AR) 4 | 5 |
| Prakash | 41 | 2018 | India | Indian | 38.2 ± 11.6 | Graft failure / functioning graft | rs699947, rs1570360, rs3025039, rs144854329 (CR) 4 | 6 |
| Gunesacar | 42 | 2007 | Germany | Caucasian | 31.7 ± 0.7 | Graft failure / functioning graft | rs3025039 (AR) 1 | 6 |
| Jimenez- Sousa | 43 | 2012 | Spain | Caucasian | 50.5 (16.6)* | CRAD / non- CRAD | rs699947 (CRAD-CR) 1 | 6 |
| Lemos | 23 | 2005 | Netherlands | Caucasian | 47.1 ± 13.5 | AR / Non-AR | rs699947, rs1570360, rs25648 (AR) 3 | 7 |
| Shahbazi | 24 | 2002 | United Kingdom | Caucasian | 39.0 ± 15.3 | RJ / NRJ | rs699947, rs1570360 (AR) 2 | 6 |
| First author | Ethnicity | AR/ CR | VEGF SNPs | Sample sizes | Statistical power | RJ | NRJ | Minor allele frequency | HWE P-value | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RJ | NRJ | Total | (α = 0.05; OR 1.5) | wt-wt | wt-var | var-var | wt-wt | wt-var | var-var | |||||||
| VEGF1 (rs699947+rs144854329) | 663 | 956 | 1,619 | 97.7 † | ||||||||||||
| 1 | Jimenez-Sousa | Caucasian | CR | rs699947 | 158 | 118 | 276 | 37.4 | 40 | 83 | 35 | 45 | 49 | 24 | 0.41 | 0.122 |
| 2 | Lemos | Caucasian | AR | rs699947 | 93 | 267 | 360 | 38.1 | 21 | 46 | 26 | 60 | 133 | 74 | 0.53 | 0.987 |
| 3 | Shahbazi | Caucasian | AR | rs699947 | 64 | 103 | 167 | 23.9 | 24 | 33 | 7 | 24 | 50 | 29 | 0.52 | 0.785 |
| 4 | Mittal | Indian | AR | rs699947 | du | du | ---- | ---- | 10 | 23 | 11 | 30 | 71 | 55 | 0.58 | 0.412 |
| 5 | Prakash5 | Indian | AR | rs699947 | 76 | 196 | 272 | 31.4 | 23 | 31 | 22 | 38 | 119 | 39 | 0.50 | 0.0027 |
| 6 | Prakash5 | Indian | AR | rs144854329 | du | du | ---- | ---- | 19 | 34 | 23 | 39 | 101 | 56 | 0.59 | 0.591 |
| 7 | Prakash8 | Indian | CR | rs699947 | 98 | 174 | 272 | 35.1 | 13 | 52 | 33 | 48 | 98 | 28 | 0.44 | 0.288 |
| 8 | Prakash8 | Indian | CR | rs144854329 | du | du | ---- | ---- | 15 | 62 | 21 | 43 | 73 | 58 | 0.54 | 0.041 |
| 9 | Prakash8 | Indian | AR | rs699947 | du | du | ---- | ---- | 48 | 98 | 28 | 13 | 52 | 33 | 0.60 | 0.288 |
| 10 | Prakash8 | Indian | AR | rs144854329 | 54 | 218 | 272 | ---- | 58 | 73 | 43 | 21 | 62 | 15 | 0.47 | 0.008 |
| VEGF2 (rs1570360) | 105 | 254 | 359 | 40.5 † | ||||||||||||
| 1 | Lemos | Caucasian | AR | rs1570360 | du | du | ---- | ---- | 47 | 38 | 8 | 118 | 119 | 30 | 0.34 | 0.999 |
| 2 | Shahbazi | Caucasian | AR | rs1570360 | 61 | 98 | 159 | 23 | 33 | 25 | 3 | 34 | 43 | 21 | 0.43 | 0.291 |
| 3 | Mittal | Indian | AR | rs1570360 | 44 | 156 | 200 | 21.5 | 13 | 16 | 15 | 48 | 51 | 57 | 0.53 | 0.00002 |
| 4 | Prakash5 | Indian | AR | rs1570360 | du | du | ---- | ---- | 27 | 31 | 18 | 35 | 115 | 46 | 0.53 | 0.013 |
| 5 | Prakash8 | Indian | CR | rs1570360 | du | du | ---- | ---- | 23 | 53 | 22 | 39 | 93 | 42 | 0.51 | 0.418 |
| VEGF3 (rs3025039) | 265 | 290 | 555 | 65.2 † | ||||||||||||
| 1 | Gunesacar | Caucasian | AR | rs3025039 | 265 | 290 | 555 | 65.1 | 231 | 31 | 3 | 230 | 55 | 5 | 0.11 | 0.423 |
| 2 | Prakash5 | Indian | AR | rs3025039 | du | du | ---- | ---- | 20 | 33 | 23 | 79 | 80 | 37 | 0.39 | 0.043 |
| 3 | Prakash8 | Indian | CR | rs3025039 | du | du | ---- | ---- | 39 | 42 | 17 | 60 | 71 | 43 | 0.45 | 0.335 |
VEGF1: vascular endothelial growth factor polymorphisms; AR: acute rejection; CR: chronic rejection; SNPs: single nucleotide polymorphisms; RJ: rejection; NRJ: non-rejection; HWE: Hardy-Weinberg Equilibrium; wt: wild-type; var: variant; du: duplicate; the 5 and 8 after Prakash indicate the last digit of publication year for these articles; values in bold indicate total sample sizes for each VEGF SNP group and significant departure from the HWE; † aggregate statistical power for the VEGF groups.
Two investigators (TE and NP) independently extracted data and arrived at a consensus. Authors of the component articles were contacted is cases of missing data. The following information were obtained from each publication: first author’s name, year of the study, country of origin, ethnicity, age of the subjects, comparators, VEGF SNPs (rs number), including transplant outcome in term of type of allograft rejection and values needed to tally the Clark-Baudouin score (Table 1). Sample sizes as well as genotype data in RJ and NRJ were also extracted along with calculated outcomes of the minor allele frequency. HWE was assessed using the application in https://ihg.gsf.de/cgi-bin/hw/hwa1.pl, HWE was reported as P-values of the controls from the Pearson's goodness-of-fit χ2-square test.
Using the G*Power program32, we evaluated statistical power. Assuming an OR of 1.5 at a genotypic risk of α = 0.05, power was considered adequate at ≥80%. Methodological quality of the included studies was assessed with the Clark-Baudouin scale33. In this scale, scores of <5, 5–6 and ≥7 represent low, moderate and high quality, respectively.
Given the hypothesis of association between VEGF SNPs and risk of allograft rejection following KT, we estimated the ORs with 95% CIs for each study by comparing RJ with NRJ among transplant recipients. Table 2 shows the frequencies of the variant (var) and wild-type alleles, as well as wt-var or heterozygous genotype (het). Non-uniformity of the variant (var) allele in VEGF1 and VEGF2 warranted the use of the allele-genotype model for VEGF1 and VEGF2. On the other hand, the var alleles in VEGF3 (rs3025039) were uniform (all < 0.50), so the standard genetic models were suitable: (i) homozygous: var–var and wt–wt genotypes compared with wt–wt; (ii) recessive: var–var versus het + wt–wt; (iii) dominant: var–var + het versus wt–wt; and (iv) codominant: var versus wt. Using raw data for frequencies, study specific risks (ORs) of allograft rejection were estimated and pooled ORs were calculated by comparing the effects on the same baseline. Multiple comparisons were corrected with the Bonferroni test. Subgrouping was based on ethnicity (Indians/Caucasians) and type of rejection (AR/CR). High significance (Pa < 0.0001) indicated strong evidence for association.
Heterogeneity in meta-analysis34 was addressed with the following: (i) its presence warranted use of the random-effects model35, otherwise fixed-effects model36 was used; (ii) estimated with the χ2-based Q test37; (iii) quantified with the I2statistic38; and (iv) sources were outlier treated. Outlier treatment divided the comparisons into pre-outlier and post-outlier.
Sensitivity analysis was used to test for robustness of the summary effects. Publication bias was considered for significant (Pa < 0.05) comparisons with ≥ 10 studies44. Significance was set at a two-sided P-value of < 0.05, except for heterogeneity estimation, which was set at Phet < 0.10)37. Data for the meta-analysis were analyzed using Review Manager 5.3 (Cochrane Collaboration, Oxford, England), SIGMASTAT 2.03, and SIGMAPLOT 11.0 (Systat Software, San Jose, CA).
Figure 1 outlines the study selection process in a flowchart following guidelines form the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA; Reporting guidelines). Table S1 (Extended data25) shows the initial search using combinations of four search strings applied to four databases resulted in 1,949 citations, followed by a series of omissions that mostly involved duplications (n = 1,924), The gray literature database (Mednar) yielded no additional papers for inclusion. Thus, the final number of included articles for this meta-analysis was seven23,24,39–43.
Of the seven articles, five23,24,39–41 examined more than one VEGF SNP (Table 1). The number of studies VEGF1 (rs699947 and rs144854329), VEGF2 (rs1570360) and VEGF3 (rs3025039) were 10, five and three, respectively (Table 2). Of the 10 VEGF1 studies, seven and three were in Indian39–41 and Caucasian23,24,43 populations, respectively. Of the five VEGF2 studies, three and two were in Indian39–41 and Caucasian23,24 populations, respectively. One Caucasian42 and two Indian40,41 studies comprised VEGF3. Table 1 shows two publications41,43 that investigated CR, which translated to three studies for VEGF1 (Table 2), otherwise, the rest focused on AR (Table 1 and Table 2).
Table 2 shows an aggregate total sample size (663 RJ/956 NRJ) and a statistical power of 97.7% for VEGF1. In contrast, both VEGF2 (105 RJ/254 NRJ) and VEGF3 (265 RJ/290 NRJ) were underpowered (40.5% and 65.2%). Mean age of the subjects was 39.96±6.6 years (± standard deviation) indicating a near to middle-age demographic profile of the KT subjects. The Clark-Baudouin scores (median 6.0, interquartile range 6.0–6.75) indicated that the methodological quality of the component studies was moderate. Control frequencies deviated from the HWE in three studies (from two articles) for VEGF140,41, two studies39,40 for VEGF2, and one study for VEGF340.
VEGF1 associations with KT. Table S2 (Extended data25) shows 32 comparisons, six of which were significant (Pa = 0.0009–0.04). Of the six, five were post-outlier derived and four survived the Bonferroni correction (Table 3). Of the four, three were in wt indicating increased risk (overall: 1.41, 95% CI 1.14-1.75, Pa = 0.002 [Figure 2], Indian: OR 1.44, 95% CI 1.13-1.84, Pa = 0.004, CR: OR 2.10, 95% CI, Pa = 0.0009) and one in var, indicating reduced risk (Indian: OR 0.61, 95% CI 0.45-0.820, Pa = 0.001). Only the CR outcome had zero heterogeneity (I2 = 0%).
| Test of association | Test of heterogeneity | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SNP group Genetic model | Comparison | Outlier status | n | OR | 95% CI | Pa | Phet | I2 (%) | Analysis model | Sensitivity outcome |
| VEGF1 | ||||||||||
| wt | Overall | Post | 9 | 1.41 | 1.14-1.75 | 0.002* | 0.17 | 31 | Fixed | Robust |
| wt | Chronic rejection | Post | 2 | 2.10 | 1.36-3.24 | 0.0009* | 0.50 | 0 | Fixed | Robust |
| var | Indian | Post | 5 | 0.61 | 0.45-0.82 | 0.001* | 0.16 | 39 | Fixed | Robust |
| wt | Indian | Pre | 7 | 1.44 | 1.13-1.84 | 0.004* | 0.16 | 35 | Fixed | Robust |
| var | Overall | Post | 7 | 0.77 | 0.60-0.99 | 0.04 | 0.14 | 37 | Fixed | Not robust |
| wt | HW-compliant | Post | 6 | 1.39 | 1.07-1.81 | 0.02 | 0.23 | 28 | Fixed | Not robust |
| VEGF2 | ||||||||||
| wt | Overall | Post | 4 | 1.58 | 1.19-2.09 | 0.001* | 0.12 | 49 | Fixed | Robust |
| wt | Overall | Pre | 5 | 1.48 | 1.01-2.15 | 0.04 | 0.09 | 51 | Random | Not robust |
| wt | HW-compliant | Post | 3 | 1.39 | 1.01-1.91 | 0.04 | 0.24 | 30 | Fixed | Not robust |
| wt | Caucasian | Post | 2 | 1.55 | 1.06-2.28 | 0.02 | 0.19 | 42 | Fixed | Not robust |
| VEGF3 | ||||||||||
| Codominant | Overall | Post | 2 | 0.69 | 0.53-0.91 | 0.01 | 0.36 | 0 | Fixed | Not robust |
| Dominant | Overall | Post | 2 | 0.66 | 0.47-0.92 | 0.01 | 0.33 | 0 | Fixed | Not robust |
VEGF: vascular endothelial growth factor gene; VEGF1: rs699947+rs144854329; VEGF2: rs1570360; VEGF3: rs3025039; wt: wild-type; var: variant; HW: Hardy-Weinberg; n: number of studies; OR: odds ratio; CI: confidence interval; Pa: P-value for association; Phet: P-value for heterogeneity; I2: measure of variability; * values in bold survived the Bonferroni correction
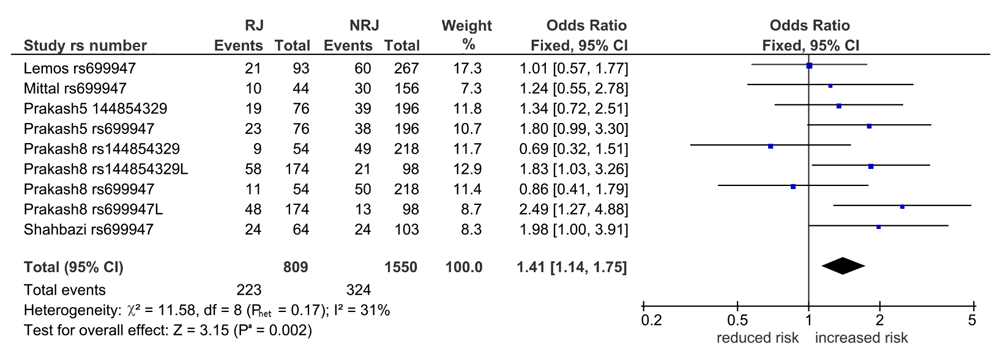
Diamond denotes the pooled odds ratio (OR) indicating increased risk (1.41). Squares indicate the OR in each study. Horizontal lines on either side of each square represent the 95% confidence intervals (CI). The Z test for overall effect shows significance (Pa = 0.002).The χ2-square test outcome has low-level heterogeneity (Phet = 0.17, I2 = 31%).
wt: wild-type; VEGF: vascular endothelial growth factor; I2: a measure of variability expressed in %; RJ: rejection; NRJ: non-rejection, L: long-term
VEGF2 associations with KT. Table S2 (Extended data25) shows 18 comparisons, four of which were significant (Pa = 0.001–0.04), were in the wt model and had moderate heterogeneity (I2 = 30%-51%). Three of the four were products of pre-outlier analysis, where the HWC outcome (OR 1.39, 95% CI 1.01-1.91, Pa = 0.04) confirmed the overall outcome (OR 1.48, 95% CI 1.01-2.15, Pa = 0.04). The other overall outcome was post-outlier derived and survived the Bonferroni correction (OR 1.58, 95% CI 1.19-2.09, Pa = 0.0001). The significant Caucasian outcome (OR 1.55, 95% CI 1.06-2.28, Pa = 0.02) contrasted with the non-significant Indian outcome (OR 1.36, 95% CI 0.72-2.58, Pa = 0.34).
VEGF3 associations with KT. Table S3 (Extended data25) shows eight comparisons, two of which were significant (Pa = 0.008–0.01) but did not withstand Bonferroni correction. These two homogeneous (I2 = 0%) pooled ORs indicated reduced risk in the dominant and codominant models (ORs 0.66–0.69, 95% CIs 0.47-0.92).
Summary of significant VEGF associations with KT. Table 3 summarizes the information on the 12 significant outcomes, five of which survived the Bonferroni correction, four in VEGF1 and one in VEGF2, all deemed robust. These outcomes identified three VEGF polymorphisms (rs699947, rs144854329 and rs1570360) that were associated with allograft rejection post-KT. VEGF1 subgroup outcomes identified CR associations and Indians to be at risk. Depending on the genetic model, the Indian population were both susceptible (wt: OR 1.44, 95% CI 1.13-1.84) and protected (var: OR 0.61, 95% CI 0.45-0.82).
The five Bonferroni-filtered findings (wt and var alleles) were either products of outlier treatment and/or subgrouping. Subgrouping identified the ethnicity and rejection type that was significant, thus specifying associations of the VEGF polymorphisms with allograft rejection post-KT. Subgrouping provided contrasts regarding significant outcomes: (i) In VEGF1, significant in Indians (Pa = 0.001–0.004), non-significant in Caucasians (Pa = 0.78–1.00); (ii) in VEGF2, significant in Caucasians (Pa = 0.02) and non-significant in Indians (Pa = 0.34); (iii) in VEFG1, significant in CR (Pa = 0.0009), non-significant in AR (Pa = 0.12). Subjecting these Pa-values to Bonferroni correction and sensitivity treatment raised the level of evidence that facilitated interpretation with greater confidence. We have shown that meta-analytical tools such as subgrouping, outlier and sensitivity treatments are instrumental in generating evidence for association. By design, such features are not present in the component single-study outcomes. This underpins the value of meta-analysis in systematically synthesizing primary study results and providing insight into associations of VEGF SNPs with allograft rejection post-KT. Conflicting outcomes between primary studies may be due to small sample sizes, hence, lack of power. Underpowered outcomes appear to be common in candidate gene studies45 and are prone to the risk of Type 1 error. In spite of the evidence for associations, the complexity of allograft rejection involves interactions between genetic and non-genetic factors allowing for the likelihood of environmental involvement. Gene-gene and gene-environment interactions have been reported to have roles in associations of other SNPs with post-KT allograft rejection. Two articles39,43 examined polymorphisms in other genes that included interleukin 18 (IL18), transforming growth factor beta 1 (TGFB1) and angiotensin II receptor type 1 (AGTR1). None of the seven articles acknowledged gene-environment interaction. Four23,39–41 of the included articles mentioned haplotype analysis with three presenting haplotype data23,39,40. Additional well-designed studies exploring other parameters would confirm or modify our results in this study and add to the extant knowledge about the association of the VEGF SNPs and renal allograft outcome.
VEGF plays a crucial role in kidney physiology with its involvement in maintaining the integrity and permeability of the glomerular capillary basement membrane17. Adaptive response of VEGF toward renal allograft tissue may be related to its angiogenic property on endothelial cells since VEGF contributes to tissue repair response of damaged capillaries23. After KT, the recipient’s neutrophils and macrophages infiltrate the allograft after reperfusion of the transplanted tissue leading to the production of VEGF24. Shahbazi et al. showed that genetically directed variations in VEGF production with increased frequency of VEGF producing alleles seemed to influence susceptibility to acute allograft rejection24. However, Lemos et al. also suggested that renal allograft recipients with genetic potential for high VEGF production had significantly better graft survival compared to recipients with low VEGF production23. Our results along the timeline of post-KT outcomes indicated increased risks, both for AR and CR in the wt allele, which agreed with Shahbazi et al.24 but contrasted with Lemos et al.23. However, the significance of our increased risk CR finding may require caution in its interpretation given the low number of studies (n = 2) and low statistical power (64.4%). More studies may be needed to clarify our CR outcome. In terms of ethnicity, Indians carriers of the wt CC genotype in rs699947 (-2578C/A), were afforded better graft survival than the CA and AA genotypes41. In contrast, Shahbazi et al. found that the -2578 C allele (rs699947) and the -1154 G allele (rs1570360) were associated with increased risk of acute renal allograft rejection in Caucasians conferring greater risk among wt homozygotes (-2578C/C and -1154 G/G) compared to -2578C/A and -1154G/A heterozygous genotypes24. These inconsistent associations among previous studies may be due to the variations in genetic background influenced by differential ethnicities of the patients.
Interpreting our findings should consider its limitations and strengths. Strengths include: (i) VEGF1 combined sample sizes translated to high aggregate statistical power (97.7%); (ii) significant HWC outcomes validated the overall pooled effects in wt. These validations served to reduce the risk of genotyping errors and minimize methodological weaknesses in our study; (iii) subgroup outcomes in CR and Indians point to potential clinical utility in the genetics of renal transplantation; (iv) efficiency of outlier treatment was the key to generating associative significance and eliminating or reducing heterogeneity and (v) stability of the core overall outcomes are underpinned by surviving the Bonferroni correction (minimizing Type 1 error risk) and robustness (determined with sensitivity treatment). On the other hand, limitations include: (i) all the component studies were underpowered; (ii) most of the moderately significant outcomes (67%) were non-robust.
To our knowledge, this is the first meta-analysis to examine associations between VEGF SNPs and risk of allograft rejection post-KT. Risks for renal allograft rejection associated with VEGF polymorphisms were shown to be increased up to 1.6-fold for the wt allele and 39% reduced for the var allele. Subgroups found to be susceptible were the Indian population and CR. These highly significant and robust core effects could render the VEGF polymorphisms useful as a prognostic biomarker in allograft rejection post-KT.
All data underlying the results are available as part of the article and no additional source data are required.
Dryad: Influence of polymorphisms in the vascular endothelial growth factor gene on allograft rejection after kidney transplantation: a meta-analysis, https://doi.org/10.5061/dryad.gqnk98skz25.
This project contains the following extended data:
Dryad: PRISMA checklist for ‘Influence of polymorphisms in the vascular endothelial growth factor gene on allograft rejection after kidney transplantation: a meta-analysis’, https://doi.org/10.5061/dryad.gqnk98skz46.
Data are available under the terms of the Creative Commons Zero "No rights reserved" data waiver (CC0 1.0 Public domain dedication).
We'd like to thank A Kunjantarachot for her invaluable contributions at the incipient stages of the draft preparation.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
References
1. Mottaghi S, Sagheb MM, Azarpira N, Abdizadeh F, et al.: Association between the Three Polymorphisms of the Glucocorticoid Receptor Gene and the Early Clinical Outcome in Kidney Transplantation Patients.Iran J Med Sci. 46 (6): 444-453 PubMed Abstract | Publisher Full TextCompeting Interests: No competing interests were disclosed.
Reviewer Expertise: Pharmacogenetics in the setting of kidney transplantation.
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Immunology, Immunogenetics, Transplantation, Molecular Biology, Virology
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 10 Feb 21 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)