Keywords
Automated data extraction, systematic review, meta-analysis, evidence synthesis, social science research, APA Journal Article Reporting Standards (JARS)
This article is included in the Artificial Intelligence and Machine Learning gateway.
This article is included in the Research on Research, Policy & Culture gateway.
This article is included in the Living Evidence collection.
This article is included in the Meta-research and Peer Review collection.
Automated data extraction, systematic review, meta-analysis, evidence synthesis, social science research, APA Journal Article Reporting Standards (JARS)
In response to feedback from all peer review reports, the following adjustments have been incorporated into the revised version of the protocol. Sections describing research objectives and methodological rationale have been modified to enhance clarity surrounding (a) objectives and scope of the proposed review, (b) study aims in light of similar efforts targeting medical research domains, and (c) identification of existing and/or emerging tools. Key items of interest have been updated to reflect potential outcomes associated with (a) benchmarking and performance assessment, (b) summary recommendations related to domain specific challenges, and (c) identification of code repositories. Revisions to search strategy and outcome reporting include additional description to better explicate (a) goals related to eligibility criteria, (b) rationale for exclusion criteria over and above inclusion criteria, (c) procedures for assessing and reporting reliability of screening and coding activities, and (d) plan for presentation of results. Adjustments to extended data include the addition of a datafile containing tabled data elements for comparison of reporting guidelines and a revised extraction techniques document incorporating a more comprehensive list of systems architectures. Figure 2 has been reformatted to enhance clarity surrounding APA guidance for applying tables/modules based on research design. As recommended by two reviewers, Figures 3 and 4 have been removed. Minor edits include word or phrasing amendments to improve accuracy and clearness.
See the authors' detailed response to the review by Sean Rife
See the authors' detailed response to the review by Frederick L. Oswald
See the authors' detailed response to the review by Michèle B. Nuijten
Across disciplines, systematic reviews and meta-analyses are integral to exploring and explaining phenomena, discovering causal inferences, and supporting evidence-based decision making. The concept of metascience represents an array of evidence synthesis approaches which support combining existing research results to summarize what is known about a specific topic (Davis et al., 2014; Gough et al., 2020). Researchers use a variety of systematic review methodologies to synthesize evidence within their domains or to integrate extant knowledge bases spanning multiple disciplines and contexts. When engaging in quantitative evidence synthesis, researchers often supplement the systematic review with meta-analysis (a principled statistical process for grouping and summarizing quantitative information reported across studies within a research domain; Shamseer et al., 2015). As technology advances, in addition to greater access to data, researchers are presented with new forms and sources of data to support evidence synthesis (Bosco et al., 2017; Ip et al., 2012; Wagner et al., 2022). An abundance of accumulated scientific evidence presents novel opportunities for translational value, yet advantages are often overshadowed by resource demands associated with locating and aggregating a continually expanding body of information. In the social sciences, the number of published systematic reviews and meta-analyses have experienced continual growth over the past 20 years, with an annual increase approximating 21% based on citation reports from Web of Science (see Figure 1).

Note. Figure was generated using the Web of Science Core Collection database. A title search was conducted in the Social Sciences Citation Index (SSCI) for articles and reviews published between 2000-2022 including variations of the terms “Systematic Review” and “Meta-analysis”. Search Syntax: ((TI=("meta-analy*" or "meta analy*" or metaanaly* or "system* review" or "literature review")) AND PY=(2000-2022)) AND DT=(Article OR Review).
Comprehensive data extraction activities associated with evidence synthesis have been described as time-consuming to the point of critically limiting the usefulness of existing approaches (Holub et al., 2021). Moreover, research indicates that it can take several years for original studies to be included in a new review due to the rapid pace of new evidence generation (Jonnalagadda et al., 2015). As such, research communities are increasingly interested in the application of automation technologies to reduce the workload associated with systematic reviews. Tsafnat et al. (2014, p. 2) delineated fifteen tasks associated with systematic reviews and meta-analyses, illuminating the automation potential for each — including the steps involved in repetitive data extraction. Recent studies and conference proceedings have outlined critical factors influencing the development and adoption of automation efforts across social and behavioral sciences. Including, but not limited to, (a) an absence of tools developed for use outside of medical science research (Marshall & Wallace, 2019); (b) a lack of universal terminology (Gough et al., 2020); and (c) nonuniformity in presenting and reporting data (Yarkoni et al., 2021). Notwithstanding these contributions, important questions related to how social scientists are addressing known challenges remain unanswered.
Social sciences encompass a broad range of research disciplines, however, what social scientists share is an interest in expanding a collective understanding of human behaviors, interactions, systems, and organizations (National Institute of Social Sciences, n.d.). Systematic reviews and meta-analyses are fundamental to supporting reproducibility and generalizability of research surrounding social and cultural aspects of human behavior, however, the process of extracting data from primary research is a labor-intensive effort, fraught with the potential for human error (see Pigott & Polanin, 2020; Yu et al., 2018). In contrast with the more defined standards that have evolved throughout the clinical research domain, within and across social sciences, substantial variation exists in research designs, reporting protocols, and even publication outlet standards (Davis et al., 2014; Short et al., 2018; Wagner et al., 2022). Notwithstanding that application of automation technologies in the social sciences could benefit from greater standardization of reporting protocols and terminology, understanding of the current state of (semi) automated extraction across these disciplines is largely speculative.
In the clinical research community, automation technologies are rapidly evolving for data extraction. Tools applying intelligent technologies for the purpose of data extraction are increasingly common for research involving Randomized Control Trials (RCT; see Schmidt et al., 2021). As data elements targeted for extraction from clinical studies and healthcare interventions often differ from those targeted by social scientists, transferability of technological solutions remains constrained. Figure 2 presents a general overview of methodologies covered by quantitative reporting guidelines applicable to social sciences per the American Psychological Association (APA, 2020). As of 2018, the APA Journal Article Reporting Standards (JARS) were updated to include clinical trial reports (represented in Figure 2 by the block labeled “Clinical Trials Module C”), incorporating elements also identified by the Cochrane Handbook for Systematic Reviews of Interventions; Higgins et al., 2022).
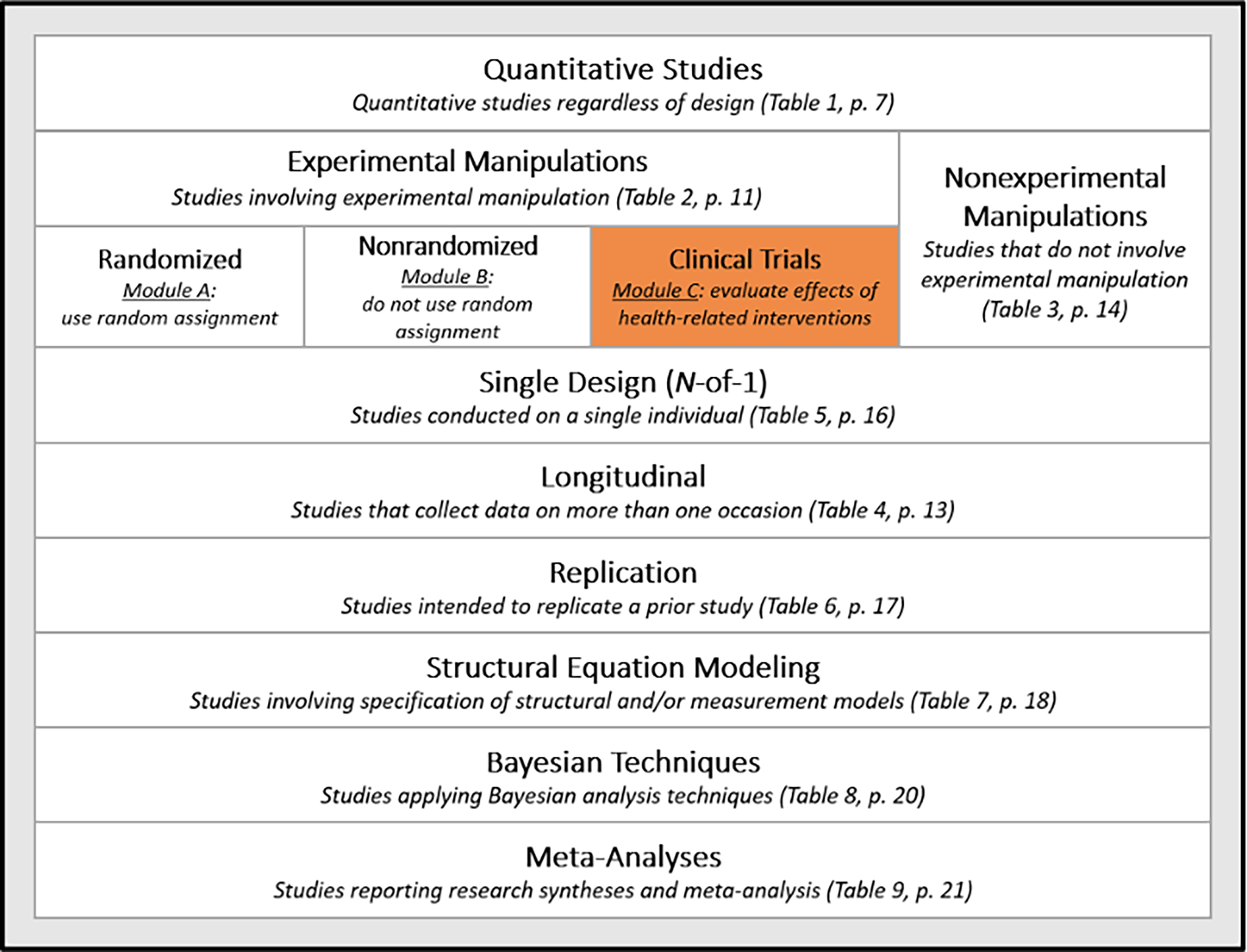
Note. Figure adapted from Appelbaum et al. (2018, Tables 1-9). APA JARS recommendations are outlined in a series of tables and modules addressing varying quantitative study designs; a singular table or combination of tables may apply to a given research report. Please note that Figure 2 includes current APA guidance released as of 2018; new tables may be added over time, please visit https://apastyle.apa.org/jars/quantitative for more details.
To elaborate, in health intervention research, targeted data elements generally include Population (or Problem), Intervention, Control, and Outcome (i.e., PICO; see Eriksen et al., 2018; Tsafnat et al., 2014). In social science research, elements targeted for extraction are similarly a function of study design, but targets can take numerous forms based on research questions considered. Researchers in social sciences often rely on APA JARS guidelines, which delineate key elements and respective reporting locations for authors to follow when presenting results of qualitative (JARS-Qual) and quantitative (JARS-Quant) research (APA, 2020; Appelbaum et al., 2018; see also Purdue Online Writing Lab, n.d.). For example, in addition to descriptive statistics (e.g., sample size, mean, standard deviation), meta-analytic efforts typically aim to extract and aggregate inferential elements such as effect sizes and p-values. Where structural equation models are involved, a researcher may be interested in extracting model fit indices; or when conducting a reliability generalization or psychometric meta-analysis (see Hunter & Schmidt, 1996), extraction of instrument psychometric properties would be imperative (Appelbaum et al., 2018; see “Extended Data” for supplementary files containing target data elements).
Given that evidence-based medicine is often associated with superior protocol standards and systematic guidelines (i.e., gold standards; Grimmer et al., 2021), the task of transferring even the most reliable automation technologies to social science research presents a substantial challenge. Even within more technical disciplines, such as Information Systems, researchers grapple with automation challenges associated with a lack of uniformity in description and presentation of constructs and measurement items (Wagner et al., 2022, p. 12). While discourse surrounding the delayed uptake of automation tools in the social sciences is occurring, the question of application transferability to domains outside of clinical research remains underexplored. Despite known barriers, delays in interdisciplinary methodological progress inhibit opportunities for collaborative knowledge synthesis both within and across fields (Gough et al., 2020). If automation techniques experiencing rapid growth in clinical research hold potential for transferability to the range of study designs prevalent throughout social and behavioral sciences, benefits could be far reaching for the development and validation of theoretical models, measurement scales, and much more.
The purpose of this study is to conduct a living systematic review (LSR) to extend the automation knowledge base by identifying existing and emergent systems for (semi) automated extraction of data used by social science researchers conducting systematic reviews and meta-analyses. We aim to uncover and present evidence that can serve as a companion project to ongoing research by Schmidt et al. (2021) who are summarizing the state of (semi)automated data extraction technologies for systematic reviews in medical research. As such, the present study holds potential to complement extant scholarship in systematic review extraction technologies by lending support for side-by-side comparison of evidence emerging from the social sciences domains with existing evidence from research medical domains. Following Schmidt et al. (2020b, 2021), who are conducting a review of data extraction techniques for healthcare interventions (i.e., RCTs, case studies, and cohort studies; see Schmidt et al., 2020a), we apply an adapted version of their methodological strategy for social science disciplines where observational research is widespread practice.1 This effort entails targeting extraction of JARS data elements identified by the APA (Appelbaum et al., 2018; see “Extended Data”).
Employing a differentiated replication framework, we apply the LSR methodology to iteratively aggregate and report: (a) the extant state of technology-assisted data extraction in social science research; (b) application trends in automation tools/techniques for extraction of data from abstracts and full text documents outside of biomedical and clinical research corpora; (c) evidence synthesis stages and tasks for which automation technologies are predominantly applied across social science disciplines; (d) specific data elements and structures targeted for automated extraction efforts by social science researchers; and (e) applied benchmarking standards for performance evaluation.
To inform this protocol and assess the extent to which our questions have been addressed in prior literature, we explored existing (semi) automated data extraction reviews. We identified six literature reviews (three static, one living, one scoping, and one cross-sectional pilot survey), two software user surveys, and one conference proceeding report. Table 1 provides a summary of scoped studies. Where some efforts focused on software applications, or “tools” that perform or assist with systematic review tasks (Harrison et al., 2020; Scott et al., 2021), others directed attention to underlying methods or techniques (e.g., machine learning algorithms) or reviewed multiple categorizations (see Blaizot et al., 2022; Schmidt et al., 2021; O’Connor et al., 2019).
| Reference | Method/Article Type (Sample Size) | Discipline(s) or Field(s) of Interest | Primary focus of project |
|---|---|---|---|
| Blaizot et al. (2022) | Systematic Review (n=12) | Health Science Research | Use of AI methods in healthcare reviews |
| Harrison et al. (2020) | Feature Analysis (n=15), User Survey (n=6) | Healthcare Research | Software tools supporting T&Ab screening for healthcare research |
| Holub et al. (2021) | Systematic Review & Cross-sectional Pilot Survey (n=78) | Clinical Trials (RCT) | Data extraction according to tabular structures |
| Jonnalagadda et al. (2015) | Systematic Review (n=26) | Biomedical Research, Clinical Trials (RCT) | Data extraction from full text; Biomedical information extraction algorithms |
| O’Connor et al. (2019) | Report/Conference Proceeding (ICASR); approx. 50 participants | Interdisciplinary | Maximizing use of technology for transfer of scientific research findings to practice |
| O’Mara-Eves et al. (2015) | Systematic Review (n=44) | Multidisciplinary (not specified) | Text mining technologies for (semi) automating citation/T&Ab screening |
| Schmidt et al. (2021) | Living Systematic Review (n=53) | Medical/Epidemiological Research, Clinical Trials | Methods/tools for (semi) automating data extraction in SR research |
| Scott et al. (2021) | User Survey (n=253) | Human Health Interventions | SR automation tool use |
| Tsafnat et al. (2014) | Scoping Review/Survey of Literature | Evidence-based medicine (RCT) | Support or automate processes of SR and/or each task of SR |
The extant knowledge base and ongoing developments surrounding systematic review automation are highly concentrated in research for evidence-based medicine (e.g., medical research, clinical trials, healthcare interventions) with limited evidence supporting how automation techniques are applied outside of the medical community (see O’Connor et al., 2019). This is not surprising given the unique relevance of systematic reviews for informing healthcare practice and policy development (Moher et al., 2015). However, while technologies to support data extraction from primary literature have advanced rapidly, many existing tools were not developed for application outside of research on the effectiveness of health-related interventions. O’Mara-Eves et al. (2015), for example, reported that text-mining techniques for classifying and prioritizing (i.e., ranking) relevant studies had undergone substantial methodological advancement, yet also highlighted that where assessment methods could be implemented with relatively high confidence in clinical research, much work was needed to determine how systems might perform in other disciplines. Other researchers similarly noted issues such as heterogeneity in testing and performance metrics (Blaizot et al., 2022; Jonnalagadda et al., 2015; Tsafnat et al., 2014) as well as risk of systemic biases resulting from inconsistent annotations in training corpora (Schmidt et al., 2021). Across projects reviewed, calls resounded for additional assessment of automation methods, including testing methods across different datasets and domains and testing the same datasets across different automation methods (Schmidt et al., 2021, O’Mara-Eves et al., 2015; O’Connor et al., 2019; Jonnalagadda et al., 2015). Despite research presenting evidence of trends toward more complete reporting (i.e., past five years; Schmidt et al., 2021), dialogue emerging from the systematic review community indicates that the time is ripe for dedicating more attention toward enhancing interdisciplinary comparability and benchmarking standards (O’Connor et al., 2019).
Existing platforms are available to support research teams in a range of time-consuming manual tasks (Blaizot et al., 2022). Even with these expediencies, not all key activities within the overall review process have received equal attention in application and technique development (O’Connor et al., 2019; Scott et al., 2021). Only a few years ago (semi) automated screening approaches such as text-mining for processing full-texts were not commonly available (O’Mara-Eves et al., 2015). As relevant study details were not always included in abstracts and often appeared throughout and across various sections of a given study (including tables and figures) discussion turned toward development of data extraction methods supporting full-text corpora (Tsafnat et al., 2014). Today, researchers supporting evidence-based medicine benefit from more robust data extraction techniques; especially efforts targeting PICO-related elements (Schmidt et al., 2021). Software tools are available for data extraction (e.g., Abstracktr, Robot Reviewer, SWIFT-Review; see Blaizot et al., 2022, p. 359), however, they have received mixed reviews related to their respective effectiveness. Notwithstanding substantial methodological strides in recent years, limited multidisciplinary reviews evaluating application effectiveness in non-clinical contexts may offer some explanation for the reported delays in uptake outside of evidence-based medicine. Further, the nominal extant research comparing techniques applied in both clinical and social contexts suggests that existing tools may not “perform as well on ‘messy’ social science datasets” (Miwa et al., 2014; as cited in O’Mara-Eves et al., 2015, p. 16). Even within structured tabular reporting contexts (i.e., tables), our understanding of technology applicability and transferability across disciplines is limited (Holub et al., 2021).
Serving as a model for the present study, Schmidt et al. (2021) reviews tools and techniques available for (semi) automated extraction of data elements pertinent to synthesizing the effects of healthcare interventions (see Higgins et al., 2022). Their noteworthy living review is exploring a range of data-mining and text classification methods for systematic reviews. The authors uncovered that early often employed approaches (e.g., rule-based extraction) gave way to classical machine-learning (e.g., naïve Bayes and support vector machine classifiers), and more recently, trends indicate increased application of deep learning architectures such as neural networks and word embeddings (for yearly trends in reported systems architectures, see Schmidt et al., 2021, p. 8). Overall, the future of automated data extraction for systematic reviews and meta-analytic research is very bright. As the earlier (i.e., preliminary) stages of the systematic review process have experienced rapid advancement in functionality and capability, development of techniques for all stages is foreseeable in the near future. Just as software tools and data extraction techniques vary in scope, purpose, and financial commitment, so too will research questions, goals, and study designs. Interdisciplinary groups and applied researchers alike call for increased collaboration to spur innovation and further advance the state of computer-assisted evidence synthesis (O’Mara-Eves et al., 2015; O’Connor et al., 2019). Though it can be inferred that not all developments spawned by the medical sciences community are easily transferrable to social sciences, necessity in fields inundated with new evidence production has carved a path for other disciplines; a path in which challenges and opportunities are openly displayed to serve as a foundation for the entire systematic review community to build upon. Additional inquiries surrounding approaches applied in social sciences may introduce previously unencountered demands that spur innovation and create valuable contributions for the entire systematic review community.
A LSR involves similar resource demands as would a static review, but is ongoing (i.e., continually reprised). The methodological rationale for selecting LSR for the proposed study is based predominantly on the pace of emerging evidence (Khamis et al., 2019). Given the uncertainty surrounding existing evidence, and the rapid pace of technological advancement, continual surveillance will allow for faster presentation of new and emergent information that may impact findings and offer value for readers (Elliott et al., 2014, 2017). Further, as this review targets published articles, the LSR methodology provides for continual search and retrieval to identify newly developed tools or techniques for which associated publications may not yet be available during previous searches. The following sections present the planned methodological approach of our living review.
This protocol is pre-registered in the Open Science Framework (OSF), an openly accessible repository facilitating the management, storage, and sharing of research processes and pertinent data files (Soderberg, 2018). This protocol adheres to the PRISMA-P guidelines (Moher et al., 2015; Shamseer et al., 2015). A completed PRISMA-P checklist is available at (Semi) Automated Approaches to Data Extraction for Systematic Reviews and Meta-Analyses in Social Sciences: A Living Review Protocol (https://osf.io/j894w). No human subjects are involved in this study.
Search strategy for this review follows existing research with protocol strategy adapted to fit goals and key elements of interest in social science domains. The model study initiated a LSR of processes supporting the (semi) automation of data extraction from research studies (e.g., clinical trials, epidemiological research; Schmidt et al., 2021, p. 26). Drawing upon the successful search strategy implemented by Schmidt et al., (2020b, 2021), we will conduct searches via the Web of Science Core Collection, IEEE Xplore Digital Library, and the DBLP Computer Science Bibliography. Databases and collections specific to clinical, medical, and biomedical literature are excluded from the search strategy (i.e., MEDLINE and PubMed). A preliminary search of Web of Science per protocol was conducted; 4,835 records were identified from Social Sciences Citation Index (SSCI), Arts & Humanities Citation Index (A&HCI), Conference Proceedings Citation Index – Social Science & Humanities (CPCI-SSH), and Emerging Sources Citation Index (ESCI). Based on the goals of the proposed study, several adjustments were made to the replicated search syntax. See “Extended Data” for relevant search strategy details, including syntax adjustments and preliminary search results. For the base review, this strategy will be replicated (to the extent possible) for remaining databases and any deviations openly reported in the project repository and subsequent publications.
The workflow structure follows existing research and guidance developed by Elliott et al. (2017) for transitioning to living status, frequency of monitoring, and incorporation of new evidence (see Figure 3). Transparent reporting of the base review and updates will follow PRISMA guidelines (Page et al., 2021). We intend to report results from the base review and later searches separately (Kahale et al., 2022). As quantity of new citations is unknown, necessary adjustments to the workflow described in this protocol will be detailed in future versions of the manuscript, noted in corresponding PRISMA reporting framework, and made available via the project repository.
The base review will begin upon publication and peer approval of this protocol. The review will be continually updated via living methodological surveys of newly published literature (Khamis et al., 2019). Updates will include search and screening of new evidence quarterly (every three months) with a cross-sectional analysis of relevant full texts at intervals of no less than twice per year (Khamis et al., 2019). Synthesis and publication of new evidence arising from continual surveillance will occur no less than once per year or until the review is no longer in living status.
Citation and abstract screening will be coordinated using Rayyan (Ouzzani et al., 2016). All citations (i.e., titles and abstracts) identified by the search(es) and all full-text documents retrieved will be screened in duplicate, with blinded and independent study selection by each researcher to reduce the risk of bias. Data extraction and coding of full text articles meeting inclusion criteria will follow the same procedure.
As screening and coding decisions required for this investigation will involve subjective judgment, the researchers will make concerted effort to strengthen transparency and replicability by conducting and reporting intercoder (i.e., interrater) reliability (IRR) assessments at multiple points throughout the study workflow (Belur et al., 2018). IRR assessment and discussion will take place immediately following completion of initial title and abstract screening, upon completion of full text screening, and again following coding of included studies. IRR assessment will be repeated for each search and update phase of the living review as detailed in Figure 3. Reliability estimates will be reported for percent agreement (ao-0.15) and Gwet’s AC1 chance-adjusted index (see Zhao et al., 2022). In the event of unresolvable disagreement(s) between researchers where consensus cannot be reached via discussion, additional qualified reviewer(s) will be consulted. All relevant details pertaining to coding decisions, resolutions and/or procedural adjustments, and underlying data will be uploaded to the project repository (see “Data Availability Statement”).
Papers considered for inclusion are refereed publications and conference proceedings in social sciences and related disciplines which describe the application of automation techniques or tools to support tasks associated with the extraction of data elements from primary research studies. As in prior reviews, English language reports, published 2005 or later will be considered for inclusion (Jonnalagadda et al., 2015; O’Mara-Eves et al., 2015; Schmidt et al., 2020b, 2021). The model article includes secondary goals related to reproducibility, transparency, and assessment of evaluation methods (Schmidt et al., 2021). The present study will also consider and synthesize reported evaluation metrics; however, we will not exclude studies omitting robust performance tests. To refine and test eligibility criteria, the complete list of Web of Science subject categories was reviewed, and inclusion decisions determined jointly by both researchers. Each category was evaluated based on the scientific branches and academic activity boundaries described by Cohen (2021). See the project supplementary data files for category selection procedures and criteria. Subjects deemed appropriate for inclusion in the initial search, title, and abstract screening stages (see Tsafnat et al., 2014) include foundational and applied formal sciences, social sciences, and social science related disciplines. Excluded subjects include natural sciences and applied clinical or medicinal science categories. In all cases, over-inclusion is prioritized to maximize search recall. See “Extended Data” for comprehensive search strategy details.
A key concern when extracting data from research corpora lies in defining the elements to be extracted; a concern equally relevant for technology-supported extraction efforts. Based on results from their ongoing review, Schmidt et al. (2021) concluded that extant data extraction literature focuses on (semi)automating the extraction of PICO elements from RCT research reports. By adapting the search strategy used by Schmidt et al. (2021), we aim to uncover discussion surrounding the application of technologies supporting data extraction from studies representing alternative research designs which do not rely on PICO reporting standards. To this end, we attempt to identify technologies that are being or have been applied across a broad range of social sciences. Our search strategy applies subject category inclusion filters to promote identification of relevant literature while mitigating potential for redundancy. The full list of included subject categories is available in the project repository (see “Extended Data”). Where an amount of overlap in extraction targets across domains is anticipated, a goal of the present study is to retrieve literature exploring the use of (semi)automated techniques which demonstrate potential for extracting APA defined reporting elements (Appelbaum et al., 2018). The eligibility criteria outlined in the following sections apply to research reports targeted for this review. Tools, technologies, and/or system architectures identified will be included in our review regardless of domain(s) of origin or domain(s) in which they are predominantly applied, given that the citing article meets eligibility criteria.
Screening decisions require exercising subjective judgment; even when coding for predetermined inclusion and exclusion of articles, decision-making can vary by coder. According to SR reliability literature, variation in coding behavior is influenced by multiple factors, including (but not limited to) subject matter expertise, academic background, research experience, and even interpersonal dynamics (Belur et al., 2018). The inclusion and exclusion criteria outlined below provide a coding framework to promote consistency, accuracy, and reproducibility in coding behavior. Variation in level of detail included in abstracts may result in a preliminary inclusion decision based on title and abstract screening and a subsequent exclusion decision based on full-text review. Where some overlap may exist across the inclusion and exclusion criteria, we elected to use a high level of specificity in developing the coding framework to facilitate detailed documentation of screening decisions for IRR assessment and reporting.
Eligible records include those which:
• employ an evidence-synthesis method (e.g., systematic reviews, psychometric meta-analysis, meta-analysis of effect sizes, etc.) and/or present a proof of concept, tool tests, or otherwise review automation technologies.
• apply an existing or proposed tool or technique for the purpose of technology-assisted data extraction from the abstracts or full-text of a literature corpus.
• report on any automated approach to data extraction (e.g., NLP, ML, TM), provided that at least one entity is extracted semi-automatically and sufficient detail is reported for:
Studies considered ineligible for inclusion in this review are those which:
• apply tools or techniques to synthesize evidence exclusively from medical, biomedical, clinical (e.g., RCTs), or natural science research.
• present guidelines, protocols, or user surveys without applying and/or testing at least one automation technique or tool.
• are labeled as editorials, briefs, or opinion pieces.
• do not apply an existing, proposed, or prototype tool or technique for the purpose of technology-assisted data extraction from the abstracts or full-text of a literature corpus (e.g., extraction of citation data only, narrative discussion that is not accompanied by application or testing).
• do not apply automation for the extraction of data from scientific literature (e.g., web scraping, electronic communications, transcripts, or alternative data sources).
O’Connor et al., (2019) described data extraction activities as the process of “extracting the relevant content data from a paper’s methods and results and the meta-data about the paper” (p. 4), therefore, we primarily target key reporting items for methods and results sections recommended by the APA. Because systematic evidence synthesis involves multiple stages, it is possible to apply multiple extraction techniques within the context of a single study and/or use a single technology to support review tasks across various stages of the same project (Blaizot et al., 2022; Jonnalagadda et al., 2015). Therefore, to support an exhaustive review and accommodate anticipated variation across automation approaches and reporting formats, a secondary area of interest includes identifying all paper sections and SR stages for which data extraction technologies have been applied (see “Extended Data”).
(Semi)automation, as defined by Marshall and Wallace (2019, p. 2) involves “using machine learning to expedite tasks, rather than complete them.” A pervasive theme throughout (semi)automation literature related to performance benchmarking is limited between-study comparability. Based on extant research, we anticipate that most included reports will incorporate basic evaluation metrics (e.g., true/false positives, true/false negatives, error) as well as other commonly reported performance measures (e.g., precision, recall, F1 scores). Though the data may prove otherwise, it is plausible that findings will mirror recent and ongoing reports revealing substantial variation in not only the type of evaluation metrics reported, but in how they are reported. For example, Schmidt et al. (2021) highlighted variety in both method and presentation of recall-precision trade off assessment (e.g., plots, cut offs, probability thresholds). They also noted that underlying algorithms represent different approaches to data extraction at the entity level, adding nuance to performance comparability (e.g., data labels, entity length, pre-classification features, training requirements). We anticipate that literature reporting tool reviews or user surveys may include measures associated with workload, such as burden, efficiency, and utility, or even more subjective assessments such user experience, cost effectiveness, or intuitiveness of software (Harrison et al., 2020; Scott et al., 2021).
Primary anticipated outcomes include identification of (a) tools/techniques applied to (semi) automate the extraction of data elements from research articles; (b) data elements targeted for extraction based on APA JARS standards; (c) systematic review and meta-analysis stages for which automation technologies are utilized; (d) evaluation metrics reported for applied automation technologies; and (e) where tools or technologies are presented, the potential for transferability across social science domains. Secondary anticipated outcomes include identification of (a) specific sections of research papers from which data is successfully extracted from primary corpora; (b) structure of content extracted using automation technologies; and (c) challenges reported by social science researchers related to the application of (semi) automated data extraction tools or technologies. Primary and secondary outcome items of interest are further described below, and supplementary data files referenced for readers to access additional information.
1. Techniques, tools, systems architectures, and/or automation approaches applied for data extraction from research documents (abstracts and full text).
2. Data elements targeted for extraction using automation technologies as outlined by JARS (APA, 2020) and further explicated by Appelbaum et al. (2018, p. 6).
3. Review tasks and stages for which extraction technologies are applied.
○ A list of fifteen tasks along with stage classifications is adapted from Tsafnat et al. (2014). See extended data files; “Review Classifications.docx”.
4. Evaluation metrics used to assess performance of the techniques or tools applied to support data extraction.
5. Transferability, availability, and accessibility of technique or tool. Target questions include:
○ Does the tool or technology easily transfer to research targeting the extraction of non-PICO prescribed elements?
○ Is the technology publicly available for use by social science researchers?
○ Where an established tool or platform was used, is it cataloged in the Systematic Review Toolbox (Marshall et al., 2022)?
○ If a technique or tool is proprietary, is it open source code?
○ Where code is open source, is it maintained in a code repository (e.g., GitHub or GitLab)?
1. Location (e.g., paper section) from which elements were extracted from research documents. To account for expected variation in reporting, paper sections of interest include, but are not limited to:
2. Structure of content from which data entities were extracted (where named).
3. Challenges identified by social science researchers when applying automation technology to support data extraction efforts. Target questions include:
○ Are there conditions under which tools are perceived as more (or less) useful than others?
○ How might technologies or tools be enhanced to better support evidence synthesis efforts across social science domains?
○ Do researchers identify reporting practices that may promote or hinder the application of automation technologies?
○ What challenges are associated with varying degrees of human involvement and/or decision-making?
Reporting of literature search and screening results will follow PRISMA guidelines (Page et al., 2021) and tailored LSR flowchart recommendations by Kahale et al. (2022). To maximize comparability with the ongoing review of extraction technologies for medical research, we plan to present the results of data extraction following Schmidt et al. (2021), who reported results in tabular, graphical, and narrative formats. Data visualization will consist of tables and figures (e.g., bar and pie charts); each new version of the review will use the same reporting and presentation structure unless new information is uncovered between published reviews that necessitates additional formats. Where appropriate, descriptive statistics will be presented in table format within each published review. Relevant corpus details will be presented in table format; where size and/or graphic requirements limit inclusion in the published review, a table containing corpus details will be maintained in the project repository and referenced in the published manuscript. We will also provide comprehensive underlying data files supporting all reported results. Underlying data files will be maintained in the project repository and updated alongside each new version of the LSR.
Authors plan to submit the base review results and subsequent update(s) to F1000Research for publication. Following the FAIR principles (i.e., findable, accessible, interoperable, and reusable; Wilkinson et al., 2016), all corresponding data will be available via the OSF project repository.
Repository: (Semi) Automated Approaches to Data Extraction for Systematic Reviews and Meta-Analyses in Social Sciences: A Living Review Protocol. https://doi.org/10.17605/OSF.IO/YWTF9 (Legate & Nimon, 2022).
Original and revised supplemental data files are available in the linked repository: Updated Supplemental Files: (Semi)Automated Approaches to Data Extraction for Systematic Reviews and Meta-Analyses in Social Sciences: A Living Review Protocol Updated. https://doi.org/10.17605/OSF.IO/EWFKP (Legate & Nimon, 2023).
This project contains the following extended data:
• Extraction Techniques Revised.docx – categories and descriptions of data extraction techniques, architecture components, and evaluation metrics of interest
• Review Classifications.docx – review tasks and stages of interest
• Target Data Elements.docx – key elements of interest for targeted data elements
• Comprehensive List of Eligible Data Elements.xlsx – comprehensive list of elements with extraction potential per APA JARS
• Search Strategy.docx – search syntax for preliminary search in Web of Science
• APA & Cochrane Data Elements.xlsx – tabled data elements for Cochrane reviews, APA Module C (clinical trials), and APA (all study designs)
Data are available under the terms of the Creative Commons Attribution 4.0 International Public License (CC-BY 4.0).
This protocol follows PRISMA-P reporting guidelines (Moher et al., 2015). Open Science Framework (OSF) Repository: (Semi) Automated Approaches to Data Extraction for Systematic Reviews and Meta-Analyses in Social Sciences: A Living Review Protocol. https://doi.org/10.17605/OSF.IO/YWTF9 (Legate, & Nimon, 2022).
1 The authors of the present study do not intend to utilize techniques developed by Schmidt et al. (2020a, 2020b, 2021) for automating search, retrieval, and relevance screening tasks associated with the LSR.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
References
1. Weissgerber TL, Milic NM, Winham SJ, Garovic VD: Beyond bar and line graphs: time for a new data presentation paradigm.PLoS Biol. 2015; 13 (4): e1002128 PubMed Abstract | Publisher Full TextCompeting Interests: No competing interests were disclosed.
Reviewer Expertise: Research methods in psychology, meta-science, automated extraction of statistical results
Competing Interests: No competing interests were disclosed.
Is the rationale for, and objectives of, the study clearly described?
Yes
Is the study design appropriate for the research question?
Yes
Are sufficient details of the methods provided to allow replication by others?
Yes
Are the datasets clearly presented in a useable and accessible format?
Not applicable
Competing Interests: I am a co-founder, employee, and shareholder of scite.ai, which uses technology related to that which is discussed in the manuscript to provide a related service.
Reviewer Expertise: Metascience; social psychology; moral and political psychology; quantitative methods
Is the rationale for, and objectives of, the study clearly described?
Yes
Is the study design appropriate for the research question?
Partly
Are sufficient details of the methods provided to allow replication by others?
Yes
Are the datasets clearly presented in a useable and accessible format?
Not applicable
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Research methods in psychology, meta-science, automated extraction of statistical results
Is the rationale for, and objectives of, the study clearly described?
Yes
Is the study design appropriate for the research question?
Partly
Are sufficient details of the methods provided to allow replication by others?
Partly
Are the datasets clearly presented in a useable and accessible format?
Not applicable
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Meta-analysis, psychometrics, individual differences, employment testing, open science
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |||
|---|---|---|---|
| 1 | 2 | 3 | |
|
Version 2 (revision) 27 Jan 23 |
read | read | |
|
Version 1 12 Sep 22 |
read | read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)