Keywords
Helicobacter pylori, Histopathology, 16S rRNA, PCR, ureA, sensitivity, specificity, Khartoum.
Helicobacter pylori, Histopathology, 16S rRNA, PCR, ureA, sensitivity, specificity, Khartoum.
Helicobacter pylori (H. pylori) is a Gram-negative, microaerophilic, spiral, and motile bacterium that colonizes the human gastric mucosa.1,2 It has been associated with the development of various clinical disorders of the upper gastrointestinal tract, such as aseptic ulcers, chronic gastritis, gastric adenocarcinoma and gastric mucosa-associated lymphoid tissue (MALT) lymphoma, which is classified as type I cancer-causing agent by the World Health Organization (WHO).3–5 Its distribution is worldwide and affects more than 90% of the world population, but it is more common in developing countries with the highest prevalence found in Africa,6,7 probably due to the possible transmission through the fecal-oral route and the unsafe sanitation conditions in these countries.1,8 Clinically, a variety of various invasive techniques (requiring endoscopy and biopsy which include, culture, histological examination, and rapid urease test, CLO (Campylobacter like organism) test, smear examination, and molecular studies) or noninvasive techniques (including serology, respiratory urea breath test, or the detection of fecal antigen) are often performed to detect H. pylori infection.9–11 The sensitivity of any of those techniques in detecting H. pylori relays on how the density of the bacterial cells within the specimens taken by biopsy (recent use of disease-related medications, specifically antibiotics and proton-pump inhibitors (PPI) can reduce the density of the cells), pathologist expertise, also the type and quality of the stain used for detection purposes.10 Many studies reported that the gold standard method for the diagnosis is the detection of H. pylori in biopsy material.12,13
Currently, many clinical laboratories use urease tests and histological analysis as a gold standard approach.13,14 In routine practice, hematoxylin and eosin (H and E), Giemsa, and immunohistochemistry staining techniques are commonly used to identify H. pylori following endoscopy; however, these techniques normally fails in identifying low numbers or coccoid forms of bacteria.15
The polymerase Chain Reaction (PCR) method offers advantages over culture and histopathology because it can detect the coccoid form of the H. pylori. PCR-based methods have been developed to detect the organism directly in clinical specimens alongside virulence and drug resistance analysis due to the high sensitivity and specificity of this technique.16 The targets of these PCR methods include the 16S rRNA gene, the urease (ureA) gene, the ureC gene, renamed phosphoglucosaminemutase (glmM), the random chromosome sequence, and the 26-kDa species-specific antigen (SSA) gene. H. pylori ureA gene is an important virulence factor that ensures that bacteria can resist acidity of the gastric mucosa.17
In Sudan, many studies were carried out to investigate the seroprevalence of H. pylori using ELISA and rapid immunochromatographic tests.18 The prevalence of H. pylori infection was estimated to be 80% among patients with gastritis symptoms, 56% with duodenal ulcer, while 60% with duodenitis and 16% apparently healthy individuals.19 In another study in Eastern Sudan high prevalence of H. pylori infection, 80% among patients with gastritis and Barrett's esophagus was reported.20 In Sudan and probably many third-world countries, the cost of diagnosis plays a major role rather than the accuracy of the diagnostic method. Hence, diagnosis of H. pylori infections is largely based on serology, detection of stool antigen and rarely endoscopy and culture. The present study aimed to compare the use of histopathology (gold standard method) with polymerase chain reaction (PCR) approach for the detection and prevalence of H. pylori infections in Khartoum State.
This was a cross-sectional study conducted at Khartoum State, Sudan between March 2018 to January 2020. The project was approved by the Ethics Committee of the Ministry of Health Research Department, Khartoum State (3/2018) (specimens were collected from patients undergoing endoscopic examination). The study aims were explained to the recruits, and a consent form was obtained and signed prior to sample collection.
Gastric tissue samples were collected by physicians from 290 patients (both gender of different age groups) undergoing endoscopic examination and suffering from dyspepsia and other gastritis-related symptoms. Patients who had received antibiotics, PPI, H2 blockers, or colloidal bismuth sulfate within the previous two months of endoscopy for treatment of gastritis or peptic ulcer, patients with a history of gastric resection, patients with complicated peptic ulcer disease, i.e. hemorrhage, were excluded.4 Two biopsy specimens were collected from the antrum and the corpus of each patient, one sample was immediately placed in tubes containing saline and transported for molecular study, while the other was fixed in 10% buffered formalin for at least 24 hours and then embedded in paraffin wax for histopathological examination.
Hematoxylin and Eosin (H and E) staining and modified Giemsa staining were performed for all samples. Three sections for each specimen were deparaffinized and hydrated in descending grades of alcohol and cut in sequential 4 μm sections. One slide was stained by routine H and E stain, and the other slide was stained by modified Giemsa stain to demonstrate the presence of H. pylori. Cover slips with DPX mounted on slides and then later examined by a histopathologist and assigned to each morphological variable.
DNA extraction of gastric biopsies was performed using the guanidine chloride method as described by Abd Al Rahem and Elhag.21 Biopsies were grounded by sterile blades and tips and then washed with phosphate buffer saline (PBS). 2 ml of lysis buffer were added, followed by 10 μl of proteinase K, 1 ml of guanidine chloride, and 300 μl of ammonium (NH4) acetate, then vortexed and incubated at 65°C for 2 hours. The mixture was cooled to room temperature, and then 2 ml of pre-cooled chloroform was applied, vortexed, and centrifuged for 5 minutes at 3000 revolutions per minute (rpm). The upper layer of the mixture was moved to a new tube, and 10 ml of absolute cold ethanol were added, shaken, and held for 2 hours or overnight at −20°C. The tube was then centrifuged for 15–20 minutes at 3000 rpm, the supernatant was carefully removed, and the tube was inverted for 5 minutes on tissue paper. The pellet was washed with 70% ethanol, centrifuged for 5 minutes at 3000 rpm. The supernatant was poured away, allowing the pellet to dry for 10 minutes. Then re-suspended into 50 μl of distilled water, briefly vortexed, and held overnight at −20°C. The extracted DNA was stored at −80°C until use.
Three different primers were used for the detection of the bacteria, targeting specific H. pylori 16S rRNA (532 bp), glmM (294 bp), and ureA (217 bp). PCR was carried out in 25 μl of reaction mixture containing 5 μl of ready to use master mix (Taq DNA polymerase, dNTPs and MgCl2) (Intron Biotechnology, Korea), 2 μl of DNA template, 1 μl of forward (F) primer, 1 μl of reverse (R) primer and 16μl distilled water (DW). For each batch of PCR assay, DW was used as negative control instead of the genomic DNA templates. The reaction mixtures were cycled in an automated thermocycler. The PCR for the specific H. pylori 16S rRNA gene was performed using the forward primer (5′-GCTAAGAGATCAGCCTATGTCC-3′) and reverse primer (5′-TGGCAATCAGCGTCAGGTAAT-3′). The PCR condition for the 16S rRNA gene was performed as follows: initial denaturation at 94°C for 3 minutes, 35 cycles of denaturation at 94°C for 30 seconds, annealing at 53°C for 30 seconds, extension at 72°C for 45 seconds, and a final extension at 72°C for 5 minutes.22 The PCR for the ureA gene of H. pylori was performed using the forward primer (5′-AACCGGATGATGTGATGGAT-3′) and reverse primer (5′-GGTCTGTCGCCAACATTTTT-3′) reported by Ye et al., which results in an amplicon of 217 bp. The PCR condition for the ureA gene was performed as follows: initial denaturation at 94°C for 3 minutes, 35 cycles of denaturation at 94°C for 30 seconds, annealing at 53°C for 30 seconds, extension at 72°C for 45 seconds, and a final extension at 72°C for 5 minutes.22
The PCR for the glmM gene was performed using the forward primer (5′-GGATAAGCTTTTAGGGGTGTTAGGGG-3′) and reverse primer (5′-GCTTACTTTCTAACACTAACGCGC-3′).23 The PCR condition for the glmM gene was performed as follows: initial denaturation at 94°C for 3 minutes, 35 cycles of denaturation at 94°C for 30 seconds, annealing at 58°C for 30 seconds, extension at 72°C for 30 seconds, and a final extension at 72°C for 3 minutes.
After amplification, 5 μl of the product was run in electrophoreses on a 1.5% agarose gel containing Ethidium bromide (0.5 μg/ml), then visualized under an ultraviolet illuminator and photographed. A 100-bp DNA ladder was used as a size marker.
Statistical analysis was done using IBM Statistical Package for Social Sciences (SPSS) software version 20.0 (RRID: SCR_019096 URL: https://www.ibm.com/products/spss-statistics). Chi-squared test was done for the analysis of categorical variables. A p-value of <0.05 was considered statistically significant.
The sociodemographic and clinical data of 290 patients recruited in this study are shown in Table 1.
Gastric biopsies were obtained from 290 patients suffering from various gastric conditions through Oesophago-Gastro-Duodenoscopy (OGD). H. pylori were clearly detected in positive samples as curved bacilli on the surface of the gastric epithelial cells; the bacteria appear as light bluish rods in H and E slides with varying sizes (3–6 μ) on the luminal surface of mucosal cells. In Giemsa’s stain H. pylori appear dark blue in a light blue background.3
From a total of 290 samples, H. pylori were found in 103 samples (35.5%). The highest number of positive H. pylori samples were observed in the active chronic gastritis followed by patients of the duodenal ulcer, gastric ulcer, and normal gastric findings in the following frequencies: 75 (25.9%), 13 (4.5%), 6 (2.1%) and 6 (2.1%) respectively, while the lowest frequency was noticed in patients with esophagitis 3 (1.0%) cases.
Among the samples analyzed by the PCR method for H. pylori 88 (30.3%) were positive using HP 16S rRNA gene, 39 (13.4%) samples were positive using glmM gene, 56 (19.3%) samples were positive using ureA gene, and the rest of samples 234 (80.7%) were negative (Figure 1).
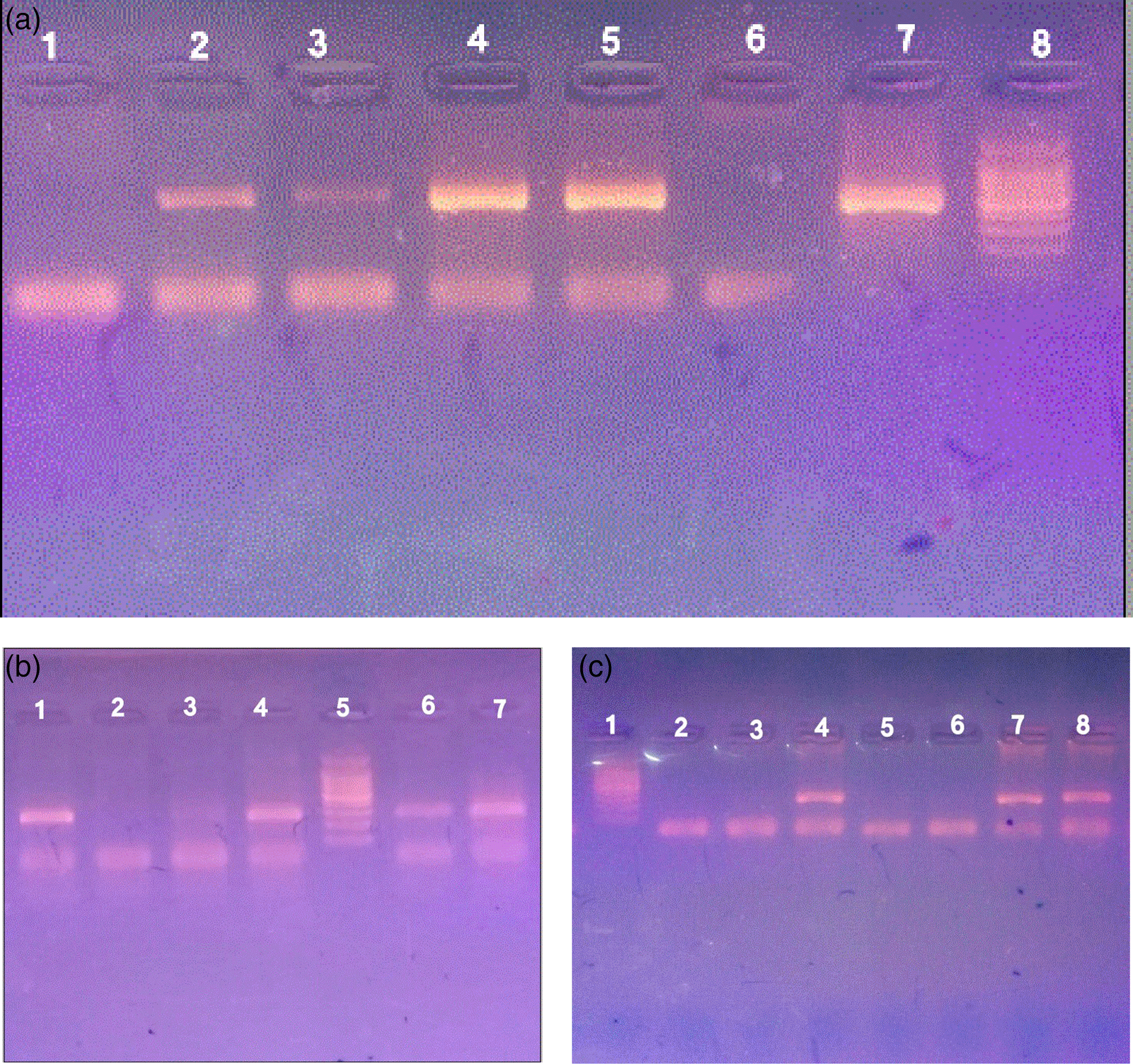
a. 16S rRNA gene. Lane 8 marker (100–1500 bp), lane 7 positive control, lanes 2–5 contain positive samples (532 bp), lanes 1 and 6 are negative samples. b. glmM gene, lane 5 marker (100–1500 bp), lane 4 positive control, lanes 1,6, and 7 contain amplicons of glmM (294 bp), lanes 2 and 3 are negative samples. c. ureA gene. Lane 1 marker (100–1500 bp), lane 8 positive control, lanes 4 and 7 contain amplicons of ureA (217 bp), lanes 2, 3, 5, and 6 are negative samples.
The calculated results were statistically significant when comparing histopathological technique results with 16S rRNA and glmM genes (p-value = 0.000) (Table 2).
Currently, there are many diagnostic methods for the diagnosis of H. pylori infections; each method has its advantages and disadvantages, so it is recommended to use at least a combination of two methods based on different principles to detect colonization by H. pylori.24 Although, the culture method is regarded as the most appropriate technique, it has limitations, particularly in case of slow-growing or fastidious bacteria, due to complicated identification and time-consuming methods. In addition to the need for immediate transport of the biopsy specimens to the designated laboratory to assure the viability of H. pylori and prevent the formation of coccoid forms of the microorganism.24–26 The histological technique and culturing of gastric biopsy specimens have been considered a gold standard method under optimal conditions.24
Histological staining enables identifying bacteria and evaluating the type and intensity of the gastric mucosa's inflammation and associated pathology, such as, atrophic gastritis (AG), intestinal metaplasia (IM), and gastric cancer or lymphoma.27
In this study, the prevalence of H. pylori infection was 35.5%. H. pylori was detected in (103/290) patients using histopathological examination with 35.5% sensitivity. There are many previous studies done in this field with various pictures of the disease. Mohamed et al. reported that 16/69 (23.2%) positive patients for H. pylori infection among Sudanese patients with colon polyps and colon cancer patients.18 Redéen et al. reported that 97/304 (31.9%) positive patients for H. pylori infection.28 In another study, Salman et al. reported that 115/210 (54.7%) samples were positive for H. pylori via histopathology, 57 (62.6%) of positive H. pylori samples were observed in patients with chronic gastritis, 11 (50%) with adenocarcinoma and 31 (44.2%) with superficial gastritis, while only one H. pylori-positive out of 5 cases observed in atrophy gastritis patient.29 Histopathology is the first diagnostic method for detection of H. pylori and is still widely used as the main diagnostic tool; nevertheless, it has limitations including higher cost, longer turnaround time, and inter-observer variation assessment; experience and skills of the pathologist do matter for the specificity and sensitivity of histopathological diagnosis of H. pylori,2 false-positive results can occur due to presence of structures similar to H. pylori24 and failure to detect all the positive samples might occur in case of intestinal metaplasia.29 The density and irregular distribution of H. pylori can vary at different sites on the gastric mucosa, which might lead to sampling error.24,27 Moreover, the sensitivity of histology may decrease in patients taking antisecretory therapy, such as, proton pump inhibitor (PPI).27
Molecular tests should be applied as replacements to the traditional method for the identification of H. pylori, which are sensitive, rapid, and precise techniques for the specific recognition of H. pylori from gastric biopsy specimens and to discover particular mutations related to antimicrobial resistance.24–26
In this study, identification of H. pylori was applied to all biopsies by PCR using specific primers. Specific H. pylori 16S rRNA gene is a conserved region of prokaryotic DNA that allows specific identification. However, H. pylori 16S rRNA gene's sensitivity and specificity were 46.6% and 78.6%, respectively. The glmM gene shows 24.3% sensitivity and 92.5% specificity. In our study, the ureA gene showed the lowest sensitivity (23.3%), and 82.3% specificity. Our result aligned with a study conducted by AlNaji et al. in 2018, which found that the glmM gene is 38.8% lower than the 16S rRNA gene 95.9%.30 Helaly et al. reported similar results (38.5%) for glmM gene.31 This low percent of glmM (ureC) gene may be due to sequence polymorphism or/in variation to the diversity of strains within the patients that reported in previous studies.30 Also, housekeeping genes are affected by geographical regions and point mutations, Intragenic and recombination are another potential factors.32
The ureA gene is a housekeeping gene that is needed for urease enzyme activity. Espinoza et al. demonstrated that the amplification of the ureA gene was noticed in (86.36%) which was lower than that of the glmM gene (100%).17 Smith et al. reported that ureA gene PCR had a very poor specificity and sensitivity.33 The possible reasons for poor sensitivity of ureA and ureC (glmM) genes for the detection of H. pylori may be that both of them are single-step PCR and thus unable to identify the lower number of bacteria or they were unable to counteract PCR inhibitors in the clinical specimens.34
The 16S rRNA gene is a useful and commonly used for the primary finding of H. pylori use Hp1, Hp2 primers with sensitivity up to 100%.30 Sugimoto et al. and Farhadkhani et al. reported that the detection of H. pylori 16S rRNA gene was greater than the ureA gene. They determined that the difference could be due to discrepancy in the primer specificity and sensitivity. Using of 16S rRNA gene for the detection of H. pylori might be more sensitive but could not be as specific as ureA gene.35,36 The poor specificity may be explained by sequence conservation across the bacterial genera and also by possible amplification of nonspecifically human DNA.34 Yet, no 100% specificity or sensitivity for primer sets amplifies H. pylori ureA and 16SrRNA genes.35,36
There is an urgent need for a rapid, accurate, sensitive, and specific test to diagnose H. pylori infections, especially on the samples collected by invasive methods (gastric biopsy). The study results suggest that H. pylori 16S rRNA gene detection by the PCR method could be used to diagnose H. pylori infections. To avoid false-positive results and increase specificity, we recommend using two conserved target genes to detect H. pylori infections.
Figshare: Underlying data for ‘Comparison of invasive histological and molecular methods in the diagnosis of Helicobacter pylori from gastric biopsies of Sudanese patients: a cross-sectional study’.
The project contains the following underlying data:
- Raw data collected from patients with gastritis symptoms: https://doi.org/10.6084/m9.figshare.17072012.v2.37
- Raw gel electrophoresis images: [PCR amplification of H. pylori on agarose gel electrophoresis 1.5%]: https://doi.org/10.6084/m9.figshare.18482015.v1.38
Data are available under the terms of the Creative Commons Attribution 4.0 International license (CC-BY 4.0).
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
Partly
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Molecular epidemiology of infectious diseases including Helicobacter pylori
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
Partly
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Partly
References
1. Pichon M, Pichard B, Barrioz T, Plouzeau C, et al.: Diagnostic Accuracy of a Noninvasive Test for Detection of Helicobacter pylori and Resistance to Clarithromycin in Stool by the Amplidiag H. pylori+ClariR Real-Time PCR Assay. Journal of Clinical Microbiology. 2020; 58 (4). Publisher Full TextCompeting Interests: No competing interests were disclosed.
Reviewer Expertise: Medical microbiology
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 2 (revision) 08 Jun 22 |
read | read |
|
Version 1 28 Jan 22 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)