Keywords
Crohn’s disease, microbiome, inflammatory bowel disease, machine learning
Crohn’s disease, microbiome, inflammatory bowel disease, machine learning
A supplemental file has been added showing the underlying code used to generate the random forest models.
See the authors' detailed response to the review by Ranko Gacesa
See the authors' detailed response to the review by Jonathan Braun
Currently, our understanding is the intestinal microbiome plays a role in the pathogenesis of inflammatory bowel disease (IBD),1 and specifically Crohn’s disease (CD).2 The RISK consortium found significant differences in the taxonomy of the mucosal and fecal microbiomes of pediatric, treatment naïve patients with CD compared to non-IBD controls.3 Similar results were demonstrated in a longitudinal study of adult patients with IBD with an emphasis on disruption of the microbiome during periods of disease activity.4
Based on an altered microbiome composition in patients with CD, microbiome signatures may be utilized as a diagnostic biomarker. From the original RISK publication,3 the addition of microbiome data to clinical information improved the performance of their classification models for CD. Similarly, Pascal et al. showed microbiome classification models for CD were accurate and performed well across 4 countries in Europe (Spain, Belgium, the UK and Germany).2
Recent data suggested that geographic bias, however, may limit the validity of microbiome based diagnostic models. He et al. studied 7,009 individuals from 1 Chinese province with 14 districts to determine regional differences in the microbiome.5 They found strong associations between microbiome composition and host district, which translated into decreased model performance when classifying metabolic diseases across districts. However, they acknowledged that other diseases, such as CD, could not be studied due to a limited sample size. Therefore, we sought to examine differences in the intestinal microbiome of pediatric patients with CD by region and to determine if geographic bias hinders the performance of a machine learning classification model across regions in North America.
A post hoc analysis of the RISK cohort was performed. The RISK cohort was a multicenter study that enrolled treatment naïve pediatric patients aged 3 to 17 years with CD and non-IBD controls from 28 sites with the United States and Canada from 2008 to 2012.3 All patients had symptoms suggestive of CD, including abdominal pain or diarrhea that prompted evaluation with a colonoscopy with biopsies from the terminal ileum and rectum. A subset of patients also provided fecal samples. Patients were either diagnosed with CD, based on endoscopic appearance and histology, or a non-inflammatory etiology for their symptoms, which served as the non-IBD controls. Full inclusion and exclusion criteria for the RISK cohort have been described in the original publication.3 In total, 447 patients with CD and 221 non-IBD controls were included in the original publication and they provided a total of 1,321 samples, including 630 ileal, 387 rectal and 304 fecal samples.
IRB approval was not required for this study, as deidentified data was used and consent was previously obtained from participants when they enrolled in the RISK cohort study.
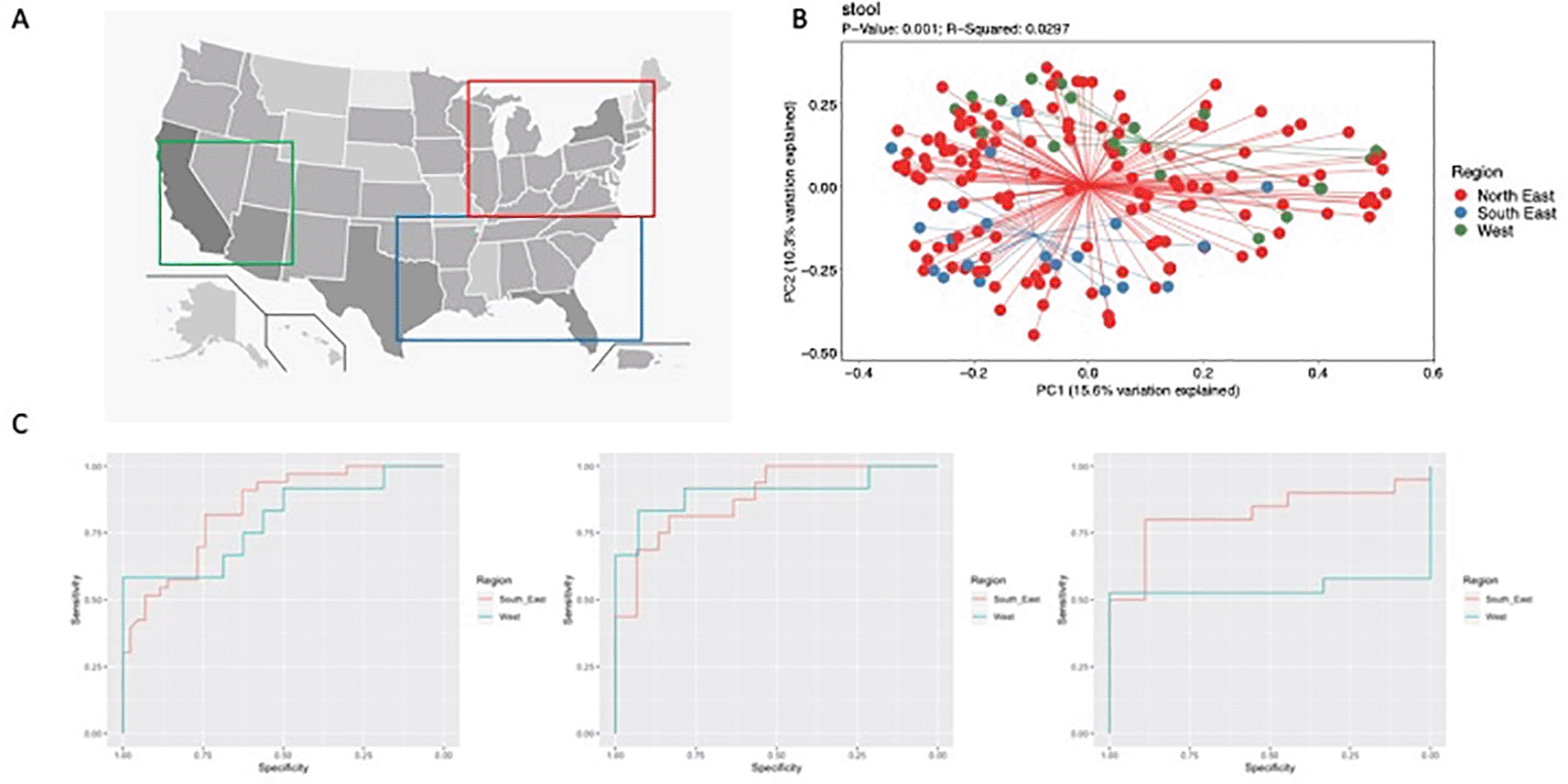
Age at diagnosis, sex, race, disease phenotype, and treatment center were examined. To evaluate the influence of geography on microbiome composition, we grouped the treatment centers into 3 subjective regions based on overall geography (North-East, South-East and West, Figure 1A).
16Sv4 rRNA gene analysis was performed in the original cohort study using the Illumina MiSeq platform. For our analysis, the original biom table was obtained and rarefied to 3,441 sequences per sample. This rarefaction depth was chosen to retain the maximum number of samples and preserve the most amount of sequencing data per sample. The alpha and beta diversity and taxonomic composition of the terminal ileum, rectum, and fecal microbiomes were evaluated using the ATIMA interface version 1.0 available through the Baylor College of Medicine Alkek Center for Metagenomics and Microbiome Research. ATIMA is a graphic user interface that allows users to provide a biom table and mapping file for microbiome analysis. To adjust for potential confounding, MaasLin was used to control for variations in age at diagnosis, sex, race, sample type and geographic region.6
Finally, we sought to develop a machine learning model to evaluate the accuracy of a microbiome model to identify patients with CD across different regions. A random forest machine learning model was trained on patients from the North-East and tested in the South-East and West using the R package healthcare.ai version 2.5.0 with the default settings. The healthcare.ai package is an open-source R package that allows for data cleaning, manipulation, imputation, tuning of models and evaluation of model performance. Visualization of model performance with AUROC metrics was done using the R package pROC version 1.18.0.
Based on the terminal ileum biopsies retained after rarefaction, we included 227 patients with CD and 165 controls with a mean age of 12.2 and 12.1 years, respectively. Approximately half of patients with CD and controls were male (58.6% and 53%, respectively) and a larger proportion of patients with CD were Caucasian compared to controls (78.9% and 68.7%, respectively). Since microbiome composition can be influenced by the presence of stricturing/fistulizing disease7 and these patients present less of a diagnostic challenge, they were excluded from our analysis to create a consistent population with an inflammatory phenotype. After separating into regions, 182 patients were in the North-East, 33 in the South-East and 12 in the West with CD, and 106 patients in the North-East, 43 in the South-East and 16 in the West without IBD.
For patients with CD, no significant differences were found in alpha and beta diversity of the ileal and rectal mucosal microbiome by geography. However, PCoA plots of unweighted and weighted beta diversity (Figure 1B) determined through the Bray Curtis metric revealed significant differences in fecal samples. In controls, no significant differences were found in alpha and beta diversity of the ileum, rectum or fecal samples. In the South-East, patients with CD had a relative increase in Fusobacteria and Bacteroidetes with a decrease in Actinobacteria and Firmicutes in fecal samples compared to the other 2 regions. This corresponded to an increase in the genera Bacteroides and Fusobacterium with a decrease in Bifidobacterium and Lactobacillus. However, after adjustment with MaasLin, Erwinia was the only genus associated with geographical variation in patients with CD. Specifically, fecal samples from CD patients in the South-East had increased abundance of Erwinia compared to other geographic regions in North America (q=0.04).
Random forest models across sample types performed well (Figure 1C, Supplement 113). The best performance occurred with ileal samples (North-East AUROC 0.89, South-East AUROC 0.85 and West AUROC 0.91). The rectal (North-East AUROC 0.87, South-East AUROC 0.83, West AUROC 0.76) and fecal (North-East AUROC 0.82, South-East AUROC 0.85, West AUROC 0.74) samples performed well, but experienced decreased performance in the West. Comparing the models, those for ileum and rectum shared OTUs discriminating CD, which included members of the Lachnospiraceae and Clostridiaceae families and the genus Blautia. Intriguingly, ileal biopsies and fecal samples shared top CD-discriminating OTUs from the Erysipelotrichaceae family and Haemophilus genus, which were not present between rectal biopsies and fecal samples.
Our results indicate that CD influences mucosal microbiome composition to a greater extent than geography in pediatric patients from North America. Machine learning classification models performed well across the regions, despite minor differences in the fecal microbiome of CD patients. Differences in microbiome composition are known to vary across populations in healthy cohorts8,9 and in patients with metabolic syndrome.5 Yatsunenko et al. showed Westernization may influence fecal microbiome composition by comparing samples from subjects in the US, Venezuela and Malawi.8 Similar patterns were seen by Pasolli et al. when they examined metagenomes from 9,428 samples from 32 countries and noted significant differences in the metagenomes of Western populations.9 Together, these studies demonstrated microbiome composition varies across populations, however, they did not address microbiome differences within countries. To that end, He et al. studied a single province in China and noted differences in microbiome composition between its districts.5 This suggested, as has been previously reviewed, that a vast number of environmental factors may play a role in shaping the microbiome and may limit the accuracy of microbiome classification models.10
Overall, our classification models performed well across regions and is consistent with prior reports. Using 2,045 fecal samples taken from patients with IBD and non-IBD controls across 4 European countries, Pascal et al. showed that a microbial signature could be used to discriminate patients with CD from non-IBD controls with an overall sensitivity of 80% and specificity of 94%.2 In a separate cohort, Franzosa et al. used metagenomics and metabolomics to distinguish IBD patients from non-IBD patients also with high accuracy.11 Our findings in pediatric CD are consistent with these results and demonstrate the feasibility of using microbiome classification models to accurately diagnose CD without geographic bias within North America.
Despite the limitations of our study, our classification models performed well. We were unable to adjust for additional confounders of microbiome composition, such as diet and supplement intake.12 However, even without this information, our models based on ileal biopsies performed well. Additionally, we noted a decrease in model performance for fecal samples and in the West, but this may be linked to a smaller sample size, which is known to hinder the performance of machine learning models. Further work with larger cohorts and different control groups will be needed to fully determine whether microbiome machine learning models can support the diagnosis of CD in children without geographical bias, and if non-invasive testing with fecal samples is feasible.
In summary, machine learning models can distinguish patients with CD from non-IBD controls without geographic bias in North America. Further development of microbiome machine learning models to diagnose CD may be warranted.
NCBI BioProject: human gut metagenome. Accession number PRJNA237362; https://identifiers.org/NCBI/bioproject:PRJNA237362.
The underlying clinical data used for this study is available through the RISK consortium. Consortium approval was required to access de-identified patient data and requests can be placed through the Crohn’s and Colitis Foundation IBD Plexus Initiative (www.crohnscolitisfoundation.org/research/granst-fellowships/ibd-plexus).
Figshare: Supplement 1. Random Forest Models. https://doi.org/10.6084/m9.figshare.21727862.v1.13
This project contains the following extended data:
Supplement 1 includes the relevant code, including used packages, inputs and outputs used to generate the random forest models. It also includes subsequent testing of the models and their outputs.
Data are available under the terms of the Creative Commons Attribution 4.0 International license (CC-BY 4.0)
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Bioinformatics, Microbiome, Artificial intelligence, Inflammatory Bowel Disease
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Immunology and microbiome in IBD
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Partly
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Partly
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Bioinformatics, Microbiome, Artificial intelligence, Inflammatory Bowel Disease
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
No
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Immunology and microbiome in IBD
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 2 (revision) 04 Jan 23 |
read | read |
|
Version 1 08 Feb 22 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)