Keywords
Colorectal cancer, 3D-sphere, primary cell culture, in silico analysis, CRC detection.
This article is included in the Oncology gateway.
Colorectal cancer, 3D-sphere, primary cell culture, in silico analysis, CRC detection.
Colorectal cancer has been found as one of the deadliest cancers worldwide and may be affected by various causes, including lifestyles, smoking habits, and western food diets. Based on the Global Cancer Observatory (GLOBOCAN), colorectal cancer is the third most common cancer with 9,000 deaths and 15,000 new cases in 2018 worldwide (Bray et al., 2018). So far, cancers have been commonly treated using anticancer, targeted chemotherapy, or particular molecular markers that are overexpressed in cancer patients (Perry & Weitzman, 2005; Kumar et al., 2017). Such progress was mainly affected by the successful breakthrough of cancer modelling, which then led to a greater connection between laboratory invention and clinical implications (Thomas et al., 2016). Therefore, cancer modelling is one of the keys to the development of therapeutic agents, while identification and validation of molecular markers are needed, respectively (Perry & Weitzman, 2005; Thomas et al., 2016).
One of the most common cancer models, in particular preclinical models, is cancer cell lines, which are pure cancer populations that are derived from cancer patients. This model has been widely used in cancer biology and also the development of anticancer drugs worldwide (Ferreira, 2013; Mirabelli, Coppola & Salvatore, 2019). Cancer cell lines were mostly well characterized in many various levels, from genetics to expression systems, including transcriptomic and proteomics characterizations (Mirabelli, Coppola & Salvatore, 2019). However, since tumors were constructed with various types of cells, cancer cell lines were lack of tumor heterogeneity such as stromal cells, and do not represent the original tumors (Ferreira, 2013; Strickaert et al., 2017). On the other hand, ex-vivo models are models that are derived from tumor tissue which may be given to animals as in vivo models or grown as in vitro models. These methods represent tumor characteristics and in vivo heterogeneity such as tumor microenvironment (Meijer et al., 2017). Moreover, ex-vivo models are not only potential for drug testing, but also models to predict patient response towards particular drugs (Meijer et al., 2017). Therefore, focusing on developing ex-vivo models is potential in many ways.
Recently, there have been several types of ex-vivo models, from in vitro to in vivo models. in vivo models may represent the tumors, for instance, using Patient-Derived Xenograft (PDX) models to predict drug responses. However, ethical issues may arise while considering using animal models in cancer research (Festing & Wilkinson, 2007; Meijer et al., 2017). On the other hand, in vitro models such as cancer cell lines do not always require ethical clearance for each experiment. Nevertheless, in vitro models lacked tumor heterogeneity, which may have later related to an unfamiliar response to the patients (Ferreira, 2013; Strickaert et al., 2017). Respectively, primary cancer cell cultures derived from patients have been acknowledged to mimic tumors in vivo and as potential models (Miserocchi et al., 2017).
Primary ex vivo models may become suitable (Miserocchi et al., 2017). However, isolation and establishment of primary cell cultures from cancer patients, in particular colorectal cancer, is found to be challenging (Failli et al., 2009). Thus, in this study, we were shown fast and simple methods to dissociate tumor biopsies for primary culture. We also identified similar expressions of three genes: ACLY, Aurora Kinase A (AURKA), and CDC20 in both ex-vivo primary cell culture models. These potential tools predict genes that are expressed while the cells grow in an in vitro environment.
The study has been approved by the Ethical Committee of Faculty of Medicine, Universitas Indonesia (Protocol ID: 20-04-0643, version 02, June 29th, 2020). Recruited subjects provided their written informed consent for biopsies donation (biobanking) and for the use and publication of their data for research purposes. All participants signed an informed consent form before participating in the study.
Biopsies were cleaned with 0.1 percent Povidone Iodine in a complete primary culture medium (DMEM) including 10% FBS and 5% antibiotic-antimycotic solutions at Biosafety Cabinet Level 2. This procedure proved crucial for successfully diagnosing primary colorectal cancer and preventing contamination by washing it with solutions containing high concentrations of antibiotics and antimycotics. Following the washing process, the biopsy was rinsed in full medium 150 mm Petri plates (Biologix, Germany) to get fragments with a diameter of 1-3 mm3. Then, fragments were pinched with sterile tweezers and scraped with a cell scraper to detach the cells from their associated tissue. After that, 5 ml complete medium was added to the fragment-containing plate and numerous times resuspended with 10 ml serological pipette tips before being passed to pre-wet 100 m cell strainers on 50 ml centrifuge tubes. Finally, the leftover pieces on the cell strain were mashed using a 12 ml syringe plunger. The medium was added to the cell top to allow the cells to flow through. After centrifuging the suspension at 300 × g for 5 minutes, the pellets were resuspended in complete mediums.
3D-sphere primary CRC was harvested on the day of 7. Approximately 10,000 cells were obtained by pipping up and down to a single cell suspension. The suspension was transferred to 1.5 ml of centrifuge tubes and spun at 300 × g for five minutes afterward. After removing the supernatant, the cells were resuspended in 100 l of DNA/RNA shield (ZymoResearch, USA). The RNA was extracted using RNA Miniprep Plus (ZymoResearch, USA) according to the manufacturer’s protocols. The RNA purity and concentrations were measured using nano spectrophotometers.
RNA was reverse transcribed to cDNA using the RT2 First-strand kit (Qiagen, Germany #Cat#330401) according to the manufacturer’s protocols. The cDNA concentration and purity were measured afterward using nano spectrophotometers.
Samples cDNA were processed to qPCR assay according to manufacturer’s instructions of RT2 Profilers (Qiagen, Hildenburg, Germany, PN.330231, Cat#PAHS-033ZC-6). qPCR was conducted using standard PCR conditions as follows: 1 cycle of 10 min, 95°C HotStart DNA Taq Polymerase is activated by this heating step, continued by 40 cycles of fluoroscence data collection in 15 s in 95°C and 1 minute in 60°C. The qPCR assay was performed in ABI 7500FAST machine (Applied Biosystem, USA). Lists of housekeeping genes were already included on the kit and the primers were already designed by Qiagen.
Protein-protein from positively expressed genes was analyzed using the String-8 Platform using multiple protein list, interactive was set to experiment, gene fusion and co-expression and highest confidence (0.9oo) developed by Jensen et al. (2009). The PPI was enriched one time by clicking “more” and represented as images.
Primary cultures were developed from two patients who had undergone surgical resections. Patients' profiles were then monitored from the Electronic Health Record of dr. Cipto Mangunkusumo Hospital, Jakarta. The CRC 001 was developed in a 57-year-old female. The tumor locations were rectal, the stadium was IV, the tumor size was T4b, N1 nodules, liver metastasis, and a well-differentiated adenocarcinoma. The CRC 002 was developed in a 45-year old male. The tumor location was the rectum. The stadium was II, N0 nodules, no metastasis, and also a well differentiated adenocarcinoma. The sample clinicopathology profiles are presented in Table 1.
Endoscopic and resected tissues were sampled, and the samples were transferred into a transport medium and delivered within 24 hours at room temperature. For resected tissues, since the samples were large, after mechanical disaggregation, viability using a trypan blue exclusion assay should be conducted to determine appropriate cell numbers that should be seeded into the well. Therefore, we seeded the samples with 10,000 cells per well to demonstrate cell morphology. Cell morphology was evaluated by visualizing the samples using an Axio Carl Zeiss inverted microscope in Axiovision Zen Blue 2.1 edition.
We grew the cells in standard media in an ultra-low u-shaped NuncSphera plate (Nunc, Thermoscientific, USA). Three–seven days after initial seeding, 3D cultures were created and captured using Carl Zeiss Microscopy, Zen Blue Editions software at 5× magnification. The majority of cells were cultured in aggregates. However, the cells were not developed into circular shapes in a regular manner. The heterogeneity of cells produced from single biopsies and their ability to develop aggregated were proven to be dependent on the biopsies' intracellular composition. Additionally, our models demonstrated significant patient characteristics, as illustrated in Figures 1 and 2.
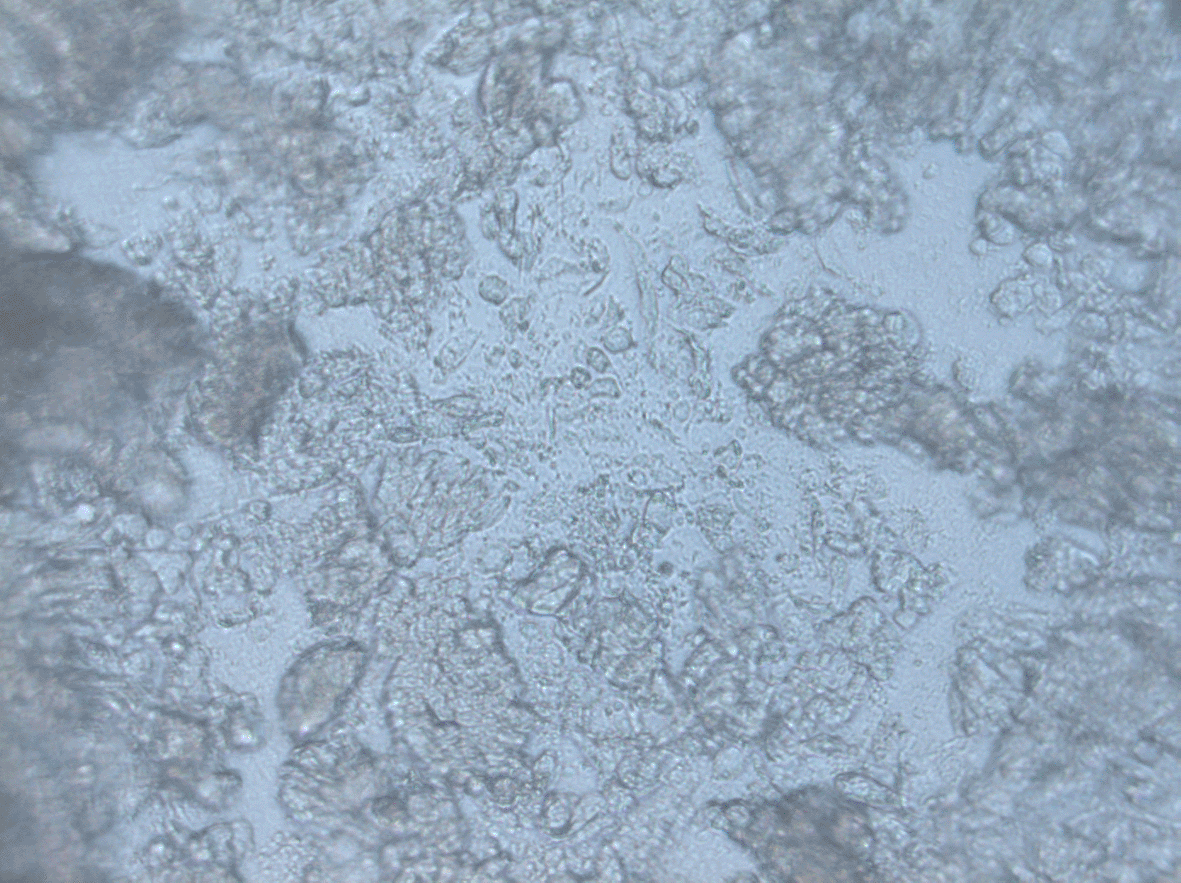
Primary cultures are 3 days after initial seeding. Cells were captured using Carl Zeiss inverted microscopy at 20 × magnification. All of the samples were derived from adenocarcinoma patients, yet distinct morphology and growth patterns were observed.
A pathway finder was used to investigate human cancer pathways, while positive expressing cells were analyzed. The CT was positioned above the signal in accordance with protocol specifications. Amplification of samples utilizing RT-Profilers using this pathway resulted in the expression of three genes, namely ACLY, AURKA, and CDC20, as indicated in Table 2. On CRC 001, the genes were largely expressed earlier than on CRC 002. However, because the data were not standardized for undiscovered housekeeping gene levels in both samples, such values were not evaluated, despite the presence of positive and reverse transcript controls. Those undetected may be due to the assay using insufficient numbers of cells (10,000 cells), which may be implied by undetected expressions. On the other hand, detected samples may be associated with elevated levels of gene expression, resulting in the detection of the genes.
There were two important pathways at this point, one involving metabolism-related genes, ACLY, and another involving cell cycle-related genes, AURKA and CDC20, respectively. As a result, this may be studied individually based on the intricacy of one pathway versus another. In Figure 3 we show that CDC20 and AURKA that has been enriched with other proteins from the STRING database, with strict criteria (highest confidence = 0.900), experiments, gene fusion, and co-expression. The co-expression score between CDC20 and AURKA was 0.808, while the experimental score was 0.487, and the combined score was 0.987 (Jensen et al., 2009). Biologically, the expression of both proteins may result in the breakdown of mitotic proteins, hence influencing chromatid segregation and mitotic protein degradation.
Ex-vivo models can be created from patients in a variety of ways, including primary culture, xenografts, and organoids (Meijer et al., 2017) Primary cell culture was certainly a supply of cells for a variety of applications, including preclinical models and personalized cancer therapy (Meijer et al., 2017; Miserocchi et al., 2017). According to our study, one of the most suitable methods for application in clinical settings was primary cell culture from patients, which may be collected concurrently with biopsies. We used samples from two distinct types of colorectal cancer biopsy in this study: endoscopic biopsies and resection tissue acquired during surgery.
One point to emphasize prior to sample collection is the use of a complete medium containing high concentrations of antibiotic and antimycotic solutions, which we did by supplementing DMEM with 10% FBS and 5% Antibiotic Antimycotic solutions. Supplemented mediums with FBS were thought to reduce cell death in these conditions, but ABAM solutions avoided contamination. Because colon tissues may have a high concentration of gut microbiotas, we recommended a beginning concentration of 5%, consistent with earlier research (Jeppesen et al., 2017; Failli et al., 2009). Using antibiotics and antimycotics at high concentrations may limit cell development, whilst using them at low concentrations below 2%, for example, 1%, increases the susceptibility of samples to be contaminated by microbial organisms.
Primary culture development can be used in a variety of ways, including two-dimensional or three-dimensional culture systems (Miserocchi et al., 2017). We employed 3D rather than 2D in our investigation. When compared to two-dimensional systems, primary culture in three dimensions results in a more physiological environment because the cells are aggregated and have three-dimensional structures that may facilitate tumor core formation and more closely imitate in vivo systems (Miserocchi et al., 2017; Riffle & Hegde, 2017). According to Riffle et al., three-dimensional tumor models were representative. In our instance, three-dimensional media will allow us to cultivate spheroids for a minimum of three to seven days without medium modifications.
We proved that growing primary cultures from resected tissue was not difficult by considering the optimal transport medium previously indicated to minimize such contamination. Samples may be extracted in around two hours using our procedures. Appropriate seeding with 10,000 cells may be employed for 3–7 days. In Figures 1 and 2, we demonstrate successful formation from two different patients (Table 1). Following that, samples can be harvested and seeded with 1,000 cells per well, as illustrated in figure 7A. However, no statistically significant variations in cell growth capacity were identified (Figure 7b), but morphological differences were observed. In the future, our simple methods may be employed to establish primary cell culture in three-dimensional (3D) systems. Additional pharmacological testing or the development of preclinical models employing 3D culture systems may be pursued in the future.
We next used the Cancer Pathway Profiler to analyze the cancer pathways that were activated in both samples utilizing this spheroid culture. The results indicated that Aurora kinase A/AURKA (NM 003600), cell division cycle protein 20 homolog/CDC20 (NM 001255), and ATP-Citrate Lyase/ACLY (NM 001096) were all expressed positively. However, housekeeping genes such as Beta actin/ACTB (NM 001101) and Glyceraldehyde-3-phosphate dehydrogenase/GAPDH (NM 002046) were not found, although positive controls and reverse transcriptase were. One possibility is that the amount of cDNA included in the samples was insufficient, resulting in the absence of multiple genes, including the housekeeping genes. On the other side, the presence of AURKA, CDC20, and ACLY may imply that these genes were expressed at a higher rate than housekeeping genes.
AURKA and CDC20 are important in cell cycle regulation in biological systems, whereas ACLY is a metabolic enzyme. AURKA is a serine-threonine kinase that catalyzes the activation of numerous proteins during mitosis (Koh et al., 2017). AURKA was a gene that induced anaphase during complex biological processes. These include the activation of spindle assembly checkpoints (SAC) and the reading of bipolar chromosome movements (Courthéoux et al., 2018). Increased AURKA expression in cancer cells may result in cells evading SAC, resulting in aneuploidy, and so inhibiting AURKA may be beneficial in future cancer treatment (Courthéoux et al., 2018). Increased AURKA expression was associated with a poor outcome in colorectal cancer liver metastases and may become a useful biomarker for prognosis prediction (Goos et al., 2013). Additionally, AURKA exhibited the potential to serve as a novel prognostic marker in colorectal cancer (Koh et al., 2017). Through co-regulation of Myc-driven oncogenes with TPX2, AURKA was demonstrated to increase carcinogenesis in colorectal tumors (Takahashi et al., 2015). Simultaneously, we generated ex vivo models from colorectal adenocarcinoma. Positive expression of AURKA may be essential for cells to develop in vitro.
Ex vivo models also expressed CDC20 similarly in our findings. These proteins are crucial for initiating anaphase by interacting with Anaphase-Promoting Complexes (APC/C). Due to the maintained CDC20 level, immature anaphase might be prevented (Mondal et al., 2007). In oral tumors, overexpression of CDC20 impairs aneuploidy (Mondal et al., 2007). TP53 and TP21 interactions negatively controlled CDC20 expression via CDH-CHE motifs in CDC20 promoters. Kidokoro et al. demonstrated that silencing CDC20 mRNA results in G2/M arrest and cell growth retardation (Kidokoro et al., 2008, p. 20). Thus, positive expression of CDC20 in our ex vivo models may demonstrate that CDC20 is required for tumor growth to continue. CDC20 and AURKA were both shown to be associated with cell cycle progression and may be related. Reactome study revealed that both genes may interact more closely during the G1-G1/S phases of the cell's growth. As a result, inhibiting AURKA and CDC20 may hold promise as a cancer treatment.
Furthermore, proteins involved in the cell cycle, such as CDC20 and AURKA we found that both cells expressed ACYL positively. ACLY genes encoded ATP-Citrate lyase, which converts citrate to acetyl-CoA and oxaloacetate (Zaidi, Swinnen, & Smans, 2012). The enzyme is required for lipogenesis and its expression was shown to be increased in colorectal cancer compared to normal mucosa. Additionally, ACLY has been implicated in medication resistance (Zhou et al., 2013). ACLY expression has been shown to decrease cytosolic citrate while increasing glycolysis. Additionally, it was observed that ACLY activates the oncogenic PI3K/MAPK kinases, leading to the Warburg effect. Additionally, Warburg may promote ACLY expression within cells via feedback regulation (Icard et al., 2020). ACLY activity generates acetyl-CoA, a precursor for fatty acid synthesis, and the Mevalonate pathway, which is required for the generation of substrates for protein prenylation, such as geranylgeranyl-pyrophosphate and farnesyl-pyrophosphate. These changes facilitate malignant transformation, invasion, and metastasis (Zaidi, Swinnen, & Smans, 2012). Additionally, by stabilizing CTNNB1, ACLY displayed greater migration and invasion. As a result, ACLY was proposed as a primary therapeutic target for both treatment and prevention (Khwairakpam et al., 2014). To summarize, employing primary ex-vivo models may help to understand how cancers grow in vitro and may be useful for predicting either the tumor's genetic profile or its application as precision medicine.
Using a mix of mechanical disaggregation and three-dimensional culture techniques, we developed preclinical ex vivo models. These techniques were convenient and straightforward to employ, allowing for the rapid production of preclinical ex vivo models in 3–7 days. In ex vivo models, the cells produced ACLY, AURKA, and CDC20, which may be linked with key genes required for cell proliferation.
Open Science Framework: Colorectal Cancer on a Dish: Exploring the 3D-sphere culture of primary colorectal cancer cells from an Indonesian perspective https://doi.org/10.17605/OSF.IO/DPYWQ (Abdullah, 2021).
This project contains the following underlying data:
‐ Amplification Plot_CRC001.jpg
‐ Amplification Plot_CRC002.jpg
‐ CRC 001.tif
‐ CT results - human cancer pathway finder.xlsx
‐ disagreggation result - CRC 001.tif
‐ Figure 1. tif (CRC 001 morphology)
‐ Figure 2. tif (CRC 002 morphology)
‐ Figure 3. png (Protein-Protein Interaction/PPI interaction of co-expressed genes)
‐ Melt Curve_CRC001.jpg
‐ Melt Curve_CRC002.jpg
Data are available under the terms of the Creative Commons Attribution 4.0 International license (CC-BY 4.0).
We thank Dr. Nadhira Nizam and Dr. Asiyah Nurul Fadilah for their valuable contribution in gathering patients and samples resources.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Partly
If applicable, is the statistical analysis and its interpretation appropriate?
I cannot comment. A qualified statistician is required.
Are all the source data underlying the results available to ensure full reproducibility?
Partly
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: antioxidant treatment
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
Partly
If applicable, is the statistical analysis and its interpretation appropriate?
No
Are all the source data underlying the results available to ensure full reproducibility?
Partly
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: tissue culture methods, in vitro models, gastrointestinal diseases, liver regeneration
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 14 Feb 22 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)