Keywords
Scientific Workflow Management, Bioimage Analyis, Usability Study
Scientific Workflow Management, Bioimage Analyis, Usability Study
In recent years, numerous workflow management systems (WMSs) for the reproducible analysis of high-throughput experimental data in life science research have been developed.1–5 It is well-established that such WMSs are indispensable to warrant transparency, reproducibility and interoperability of increasingly complex data analysis tasks that typically combine a number of different data analysis tools and subtasks. Matching the needs of different user groups and application settings, WMSs have been implemented as either graphical user interfaces1–4 or as script-based tools,5 addressing a broad variety of user groups and an equally broad range of needs for user interaction or operability on large data sets.
Many of the prominent WMSs have been developed and maintained over many years and are now mature software systems whose relevance and popularity are undermined by large citation numbers. However, apparently little attention has been spent on systematically and formally exploring workflow management from the viewpoint of usability. Yet, usability is of interest in many respects. First, a detailed investigation of software usability can guide future developments of specific tools, and more generally may help to summarize experiences to formulate guidelines for future developments.6,7 Second, the ergonomics of workflows affect the outcome of data-centric research studies. Since many studies and corresponding data analysis workflows involve a human-in-the-loop,8 usability aspects inevitably determine the inductive bias of obtaining insights from data. A thorough investigation of usability is thus not just a matter of assessing convenience and efficiency, but also a matter of elucidating the role of software in the scientific method.
In this contribution, the results of a formative usability study in the context of label-free digital pathology9 is presented. The goal of this study is to provide a first case study on how usability studies for scientific workflow management can be conducted, and how they can systematically be used for improving WSMs. Specifically, a study that investigates and compares two distinct groups of users following two different workflows was conducted to solve one and the same image analysis task.
This study deals with a setting in label-free digital pathology based on hyperspectral infrared microscopic imaging data and it is based on a workflow originally established by Kallenbach-Thieltges et al. 2013,9 which was the basis for a number of applications of infrared microscopy in different clinical settings.10–12 In label-free digital pathology, tissue samples are measured using an infrared microscope. The infrared microscope yields a hyperspectral microscopic image with a spatial resolution of around 5 μm, where each pixel is represented by an infrared spectrum covering a wavelength bandwidth of several hundred optical channels. To identify disease-associated tumor regions in tissue samples, the images need to be pre-segmented using unsupervised learning approaches which allows to extract training data for a supervised classifier, as illustrated in Figure 1.
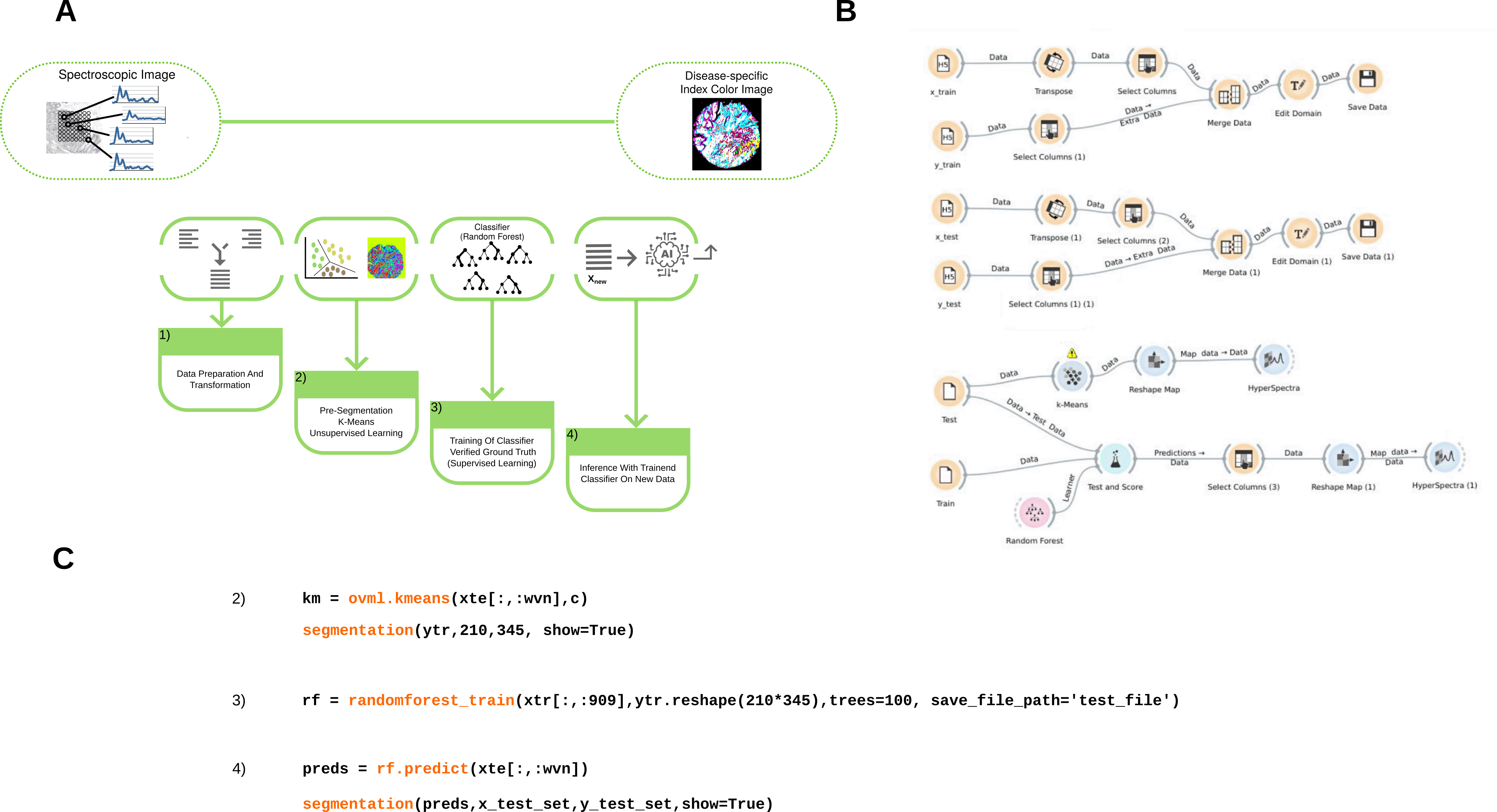
Panel A) shows the workflow as abstract scheme, while panel B) shows the same workflow implemented in the Orange framework. Panel C) gives an impression of the main implementing this workflow in OpenVibSpec.
This study involves two different implementations of this workflow. First, a script-based implementation from the OpenVibSpec tools was used,13 which constitutes a Python re-implementation of the workflow used in previous studies.9–12 The second implementation was based on the graphical user interface (GUI) of the Orange WMS.1 Furthermore, two distinct user groups with different levels of programming skills and life science background were recruited to investigate their usage patterns across the two different implementations. In addition, this study addresses the question of how to evaluate a given workflow for biomedical image analysis.
As documented by the International Standards Organisations (ISO) in ISO 9241, the usability of a product or a system is defined as the extent to which a user can use the system to achieve his/her goals “effectively, efficiently and with satisfaction”.14 In its core, usability is not one single factor of a system or product but multiple factors of the system or product contribute to or reduce its usability. These factors or concepts are: learnability, efficiency, memorability, errors and satisfaction.6 Usability is seen as relevant during the whole lifetime of a product or system, from development to procurement, and is an important aspect of the perceived user experience (UX) of a product or system.
To evaluate the usability of a system, a variety of methods and types of usability tests or inspections can be used. Such evaluations can differ, for example, in the participants’ knowledge (expert vs. novice user), in its test design (task-based test vs. cognitive walkthrough), or if it is done during the development or with a final version of the system (formative vs. summative evaluation).6,15,16 Besides test-based evaluations, heuristic usability inspections are also commonly used.17
Formative studies are of a rather exploratory nature and can be conducted very early in the development phase of systems, even on the basis of simple wireframes or prototypes to define requirements and reflect on the interaction design at an early design stage.16 In these studies, mainly qualitative data is collected, for example comments and impressions by users regarding the interaction design of the system, if and how successful a task could be completed or how they perceived the systems feedback (cf. Ref. 15). Common methods for formative usability studies are, for instance, thinking aloud tests, in which users are encouraged to articulate their thoughts during use or task-based tests with follow-up interviews. In summative studies, a more formal approach is taken and attempts are made to demonstrate the effectiveness of a newly developed system. To conduct a summative usability test, at least functioning prototypes are needed. A typical goal of a summative test would be to highlight the benefits of a new system or interface to other existing ones. However, the boundaries of formative and summative testing are not rigid, as a summative test could also be applied after several development stages of a system.
To assess the usability of a system, a combination of different metrics and methods can be applied to collect the data of interest. However, the type of test can affect the number of participants in a study. For example, an expert-based usability evaluation can possibly be applied with a smaller number of participants than for example user-based usability tests. Yet, the number of needed participants in usability tests is under dispute.18–22
In this study, a formative approach for the usability evaluation was used. The primary aim of this study is not to compare the efficiency of both tools but rather to explore possibilities and constraints designing usability studies for scientific workflow management tools and how such usability studies contribute to their improvement.
In order to generate a broad spectrum of insights and perspectives, a formative approach was chosen for the usability evaluation of the two WMS. A task-based study with participants was conducted, in which participants were asked to use a combination of supervised and unsupervised machine learning algorithms to classify microscopic data of tissue images, as shown in Figure 1. During use, observations were noted by the study facilitator and participants were asked to express thoughts aloud. After completing the tasks, a semi-structured interview was conducted with each participant to verify the observations made and to solicit further perspectives. The interview incorporated questions based on Nielsen’s usability heuristics.17
Two alternative implementations of the workflow were investigated, one that could be used via script and one that offered a GUI. For the script-based implementation of the workflow, the Python-based implementation provided in the OpenVibSpec tools13 was utilized with a Jupyter notebook23 as an interface (See source data).24 The implementation provides structured access to perform clustering of image spectra, to extract training spectra from annotation masks, and to train and validate supervised classifiers. Following common practice,9 -means clustering25 was employed for unsupervised segmentation combined with random forests26 for supervised classification.
The study GUI based implementation relies on the functions provided by the Orange framework1 (See source data).24 In Orange, a workflow can be created by connecting different widgets. These widgets represent certain data operations on a data set and usually allow some kind of configuration. As illustrated in Figure 1, the data for the workflow was pre-established in the interactive Orange GUI, as data import and export operations were already added. Thus, users were only required to create the workflow with suitable data operations and adjust parameters. This adaptation of the workflow saved considerable time for the study, as the import and export operations demanded a noticeable amount of time. The final workflow is illustrated in Figure 1A.
In order to facilitate at least basic comparison between the two implementations, this study was limited to those methods that are implemented in both platforms, noting that both Orange and OpenVibSpec are based on the same standard libraries, most notably Scikit-learn27 and Numpy.28
For both implementations, participants were briefed with a targeted training video to explain the technical realization of the workflow, in order to warrant the necessary level of prior knowledge for the study (See source data).24 Participants had the chance to watch the video or parts of the video again at any time during the study.
The data set was derived from previously published studies investigating colorectal carcinoma using infrared microscopy.11,29 In brief, the complete data set comprises infrared microscopic images of roughly 200 tissue-microarray (TMA) spots of more than 100 different patients, where each spot is roughly 1 mm in diameter and represented by an infrared microscopic image covering roughly 250×250 pixel spectra. TMA slides were purchased from US Biomax Inc., MD, USA, and the infrared microscopic images are accompanied by conventional hematoxylin and eosin (H&E) stained images which were acquired subsequent to infrared microscopy. For the script-based implementations, many images of a complete TMA spot were used as a basis for the study. For the GUI-based implementation, images were reduced in size to 224×224 pixels for the training data and 195×210 for the test data in order to meet constraints in terms of computational resources and time window per participant.
For the study, 22 participants (7 females, 15 males) were recruited in total, among students of two different study programs at the Ruhr-University Bochum with ages ranging from 23 to 39 years (M = 26.05, SD = 3.7) (see Underlying data30).
In order to take account of the study’s objectives in the selection of test users, two target groups were deliberately sought which correspond to the users of the tools compared, namely Orange and OpenVibSpec.
The first group was recruited among students with at least advanced undergraduate knowledge in a life science study program and no or limited programming skills (6 females, 5 males, mean age = 25.27, SD = 2.83). The second group was recruited among computer science students at comparable level, possessing good programming skills, but no or limited background in the life sciences (1 female, 10 males, mean age = 26.82, SD = 4.26).
To counteract a bias of the study by specifically selected test persons, all participants were deliberately selected in such a way that they did not have any special expertise in the actual question being compared, i.e. microspectroscopic tissue characterisation. Based on this, all participants were provided with the same training material, which should shed more light on the background of the data origin and the methods used. Care was taken to ensure a lightweight introduction was given to enable all participants to concentrate fully on the task during the actual test series.
The implementations were made available to participants on a desktop computer equipped with an Intel i7-2600 CPU at 3.40Ghz and 8 GBytes memory. The actual implementations were installed on a remote Linux server with 64 CPUs and 512 GBytes of main memory, and made available on the desktop computer through a remote desktop connection, which was also equipped with a Linux OS.
The task had to be adapted in such a way that it could be carried out with both tools, but at the same time produce a result that could be interpreted in the sense of the introductory material provided to the participants; This last requirement and the general orientation of the two tools caused a discrepancy in the execution speed of the Orange tool, which is describe in more detail in the results section. Since the data reading and saving processes also took several minutes, they were skipped in the task for Orange and the participants were given a minimally prepared workflow in which the training and test data widgets were already provided.
The procedure for each participant was conducted as follows: First, each participant completed a demographic questionnaire and gave their consent to participate in this study (see Underlying data30). Then, participants watched an introduction video with educational material on workflow-based hyperspectral infrared microscopic imaging data analysis. After watching the video, participants started the task with either the GUI tool or the script-based implementation. The instructions to complete the script-based workflow were given in the Jupyter environment.31 For the GUI workflow, participants received the instructions on how to complete the task on paper and PDF. During the study, participants had the chance to ask questions, e.g. if they got stuck or had any other kind of problem to complete the task. While using the tools, the participants were also asked to think out loud. During the task, their screen was recorded and notable usability issues were documented by the study facilitator. After completing both workflows, a semi-structured interview was conducted with each participant in which the usability of both tools was focused.
This semi-structured interview consisted of general questions about the participants and their prior knowledge, as well as questions integrating Nielsen’s usability heuristics17 (see Underlying data30). The interviews made it possible to specifically question observations that we had documented with the respective participants and to judge their comments and wishes more accurately.
The results are structured into three subsections. First, is the specific task-related observations and results gained from participants’ use of both tools. In the following several usability problems are provided with suggestions for improvement. All the usability issues that occurred were not listed, however the examples that had a major negative impact on task completion in this study were mentioned. Lastly, task completion and task understanding in each of the two participant groups were examined, and how user studies on real data can be conducted to help make WMS available to a wider range of users was analysed.
Task completion
Regarding the task completion time, the group of life science students needed slightly longer to finish the task in OpenVibSpec and, on average, around 5 min longer to finish the task in Orange (see Table 1) (see Underlying data).30 The task completion time did not differ notably between Orange and OpenVibSpec for the computer scientists. The time differences for the life science group were on average around 3 min. These results suggest two aspects. First, the task duration was reasonably evenly distributed between the two systems, as both groups needed around the same time to finish the task. Second, the results also show that the computer scientists finished the task faster with both tools in this study. It should be noted, however, that participants were not asked to finish the task as quickly as possible, but could do it on their own pace. Therefore, the faster completion time of the computer scientist group can probably be justified by a more practiced use of computers in general.
| Group | Orange | OpenVibSpec | ||
|---|---|---|---|---|
| M | SD | M | SD | |
| Life Science | 39:44 | 08:23 | 36:33 | 07:56 |
| Computer Science | 34:05* | 06:43 | 33:44 | 06:05 |
Task complexity
The task was rated as quite complex by the life scientist group. Using the scripted approach with OpenVibSpec, all participants managed to complete the task successfully. However, 10 out of 11 participants of the life sciences group needed very close supervision and a fair amount of assistance, as the learning curve seemed too steep without prior programming knowledge. For example, exchanging a simple placeholder variable with a concrete integer value for the segmentation of the image was not obvious to many, although it was clearly described in the instructions. Without the intervention of the study facilitator, some participants would have already given up on this task at this point. Questioned afterwards, the participants also stated that the instructions actually clearly described the exchange of the placeholder variable for an integer value. Some said they were simply overwhelmed by the task and by the code. As for the computer science group, still 5 out of 11 participants needed some sort of support during the script-based task. Overall, interviews indicated that the task was easier to solve with the GUI. Especially participants who found the script-based variant somewhat discouraging positively emphasized the GUI, as the GUI enabled them to follow the workflow at least visually and, despite the instructions, they had the feeling of being in control. However, even with step-by-step instructions for the GUI, it was a highly challenging task for novice users as most of them relied on support by the study facilitator at one point or the other. In conclusion, task complexity can be interpreted in several ways. The different perception of the two approaches may have been due to the differently prepared instructions (once text instructions in Jupyter, once screenshots for Orange). However, this is contradicted by the fact that at least one participant in the Life Science Group was able to solve the task who had neither Python nor programming experience. Instead, the perception of complexity seems to be based more on programming skills in general. Many participants, especially in the life science group, had to learn how to run programming code in the Jupyter environment at the beginning of the task. Some participants even had difficulties recognizing which sections belonged to the instructions and which belonged to the script on their own, until they received support from the study facilitator. Such difficulties made the barrier to solve the task even higher for these participants.
Comprehension of results
Participants’ understanding of the workflow outcomes was quite heterogeneous. Although all participants were given a basic introductory video to watch before the first task, four life scientists and four computer scientists were unable to give any real conclusion about the outcome images, such as what the classification of the images could mean and how one would proceed further. In total, four participants (two Computer Scientists, six Life Scientists) expressed that they had followed the steps of the instructions but could not elaborate on the context of the subject matter. Overall, it had been easier to understand the workflow in Orange, since the widget names would have at least made it understandable that a data set was manipulated with several operations. The majority of participants (nine computer scientists, five life scientists) were able to explain that the workflow applied artificial intelligence methods to classify tissue data. Furthermore, they were also aware that they could not interpret the results in more detail and would have to present the result to an expert to repeat the clusterings with other parameters if necessary. This shows that although the participants did not know the dataset before participating in the study, nor had they used the tools before, they were able to perform the task and also were able to comprehend it well in terms of its subject matter. In addition, one participant explicitly pointed out that the resulting images lacked a unique assignment of index colors. It was impossible to understand how the method decided which areas of the image were clustered together. Here, the participant mentioned the use of explanatory labels to understand the clustering.
Severe usability issues
The usability issues presented here refer primarily to those that made designing the task for the study challenging and those that hindered or negatively impacted participants’ task completion.
Learnability
As mentioned in the previous section, participants of the life science group had severe problems in completing the task on their own with the script-based approach of OpenVibSpec. The learning curve seemed too steep for the majority of these participants despite detailed instructions. However, the learnability of OpenVibSpec was disrupted in that many participants first had to learn how to use the Jupyter notebook, e.g. to run scripts or even to understand the controls at all. The task became considerably easier if someone had previous experience in programming. Participants with prior experience in Python and Jupyter notebooks had little to no problems focusing on and solving the task. In comparison, all participants were able to replicate the workflow independently with the GUI of Orange, even though none of the participants knew the application beforehand.
Efficiency
Prior to the usability test, during the preparation of the study, longer computing times for Orange was recognized. Therefore, the data set used in this study was reduced for Orange. Since the data reading and saving processes also took noticeably longer computing times, they were skipped in the task for Orange, and the participants were given a minimally prepared workflow beforehand. During the interviews, several participants from both groups presumed that the tools share the same code base, except that one offers a GUI and the other only script-based. These assumptions made by the participants suggest that the changes to the workflow (smaller data set for Orange) and to the task (skipping the import and export operations in Orange) resulted in both tools being considered equivalent in terms of efficiency and considered comparable.
In addition, it can be noted that participants who had no programming experience rated the efficiency of Orange better. The reason given was learnability: In OpenVibSpec, a longer training period would be necessary and programming experience would be required, whereas Orange could be operated directly via the GUI.
Error messages and error prevention
An important usability criterion is the handling of error messages, which should represent the error in a human-readable form. During the study, error messages occurred in both tools that were basically Python error messages and could only be interpreted by participants with programming experience. For all other participants, the study facilitator had to intervene and provide assistance to the participants. In addition to this, in the Orange GUI it was not always clear to the participants when an error message was displayed and when it was only a warning message. The occurrence of such error and warning messages led to uncertainty in many participants’ completion of the task, which is why the study facilitator explained the messages shortly and participants could continue with the task.
Another usability issue concerns error prevention in general. An example for a missing error prevention that hindered participants to achieve the expected resulting image occurred during the classification with the random forest algorithm in Orange. Only 7 out of 11 participants from the computer science group were able to successfully complete the task. Three achieved a different result image via incorrectly connected widgets. In the life sciences group, 6 out of 11 participants were able to correctly solve the task with Orange. For 5 out of 11, the same connection error of two widgets occurred. This error was a result of a rather simple usability issue while creating the workflow. However, users did not realize they had done something wrong until they saw the resulting image, as the feedback given by the GUI was not explicit and clear enough. These participants did not notice that the order in which they connected the training and test data to a specific widget led to an automatic classification of the data by the tool as training or test data. Due to the incorrect order of the connections, training data was labeled as test data and vice versa (see Figure 2). Here, the feedback to the users was not clear enough, as it was not noticed by those to whom it happened. On the other hand, this error could have been prevented if the order of the connection had not mattered. In OpenVibSpec such an error did not occur because an automatic interpretation of the dataset is not offered, but one had to explicitly specify the training dataset or test dataset in the method call.
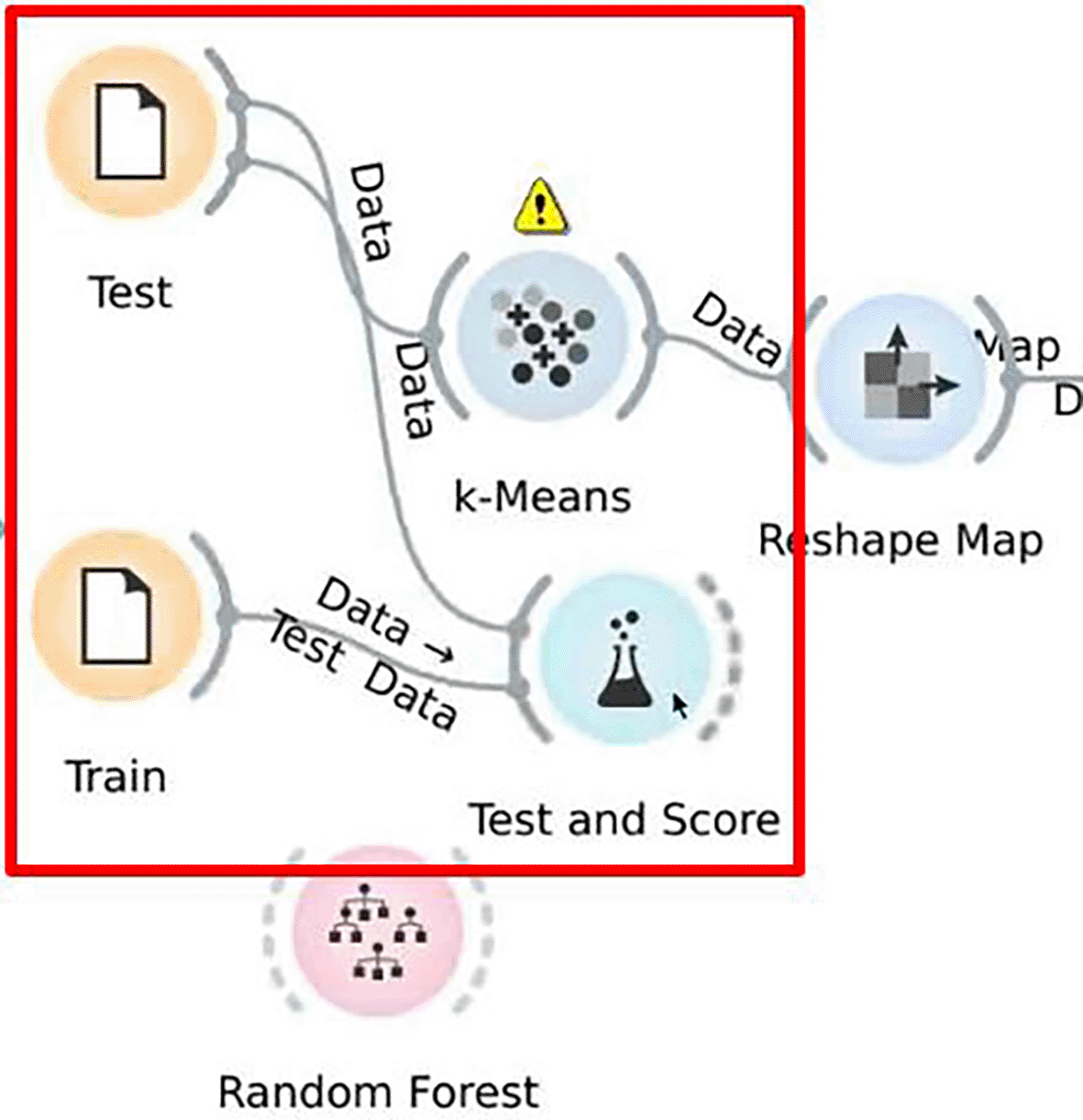
If afterwards the the training data (‘Train’) was connected with ‘Test and Score’, the tool labeled the training data as test data. This issue occurred to 8 participants in total.
The data revealed that the participation of two different user groups resulted in a broader range of views, task-related observations, and usability problems found.
For example, according to the statements in the interview and the observations during the study, it was apparent that most participants had a more pleasant user experience with the GUI tool, in particular in the life scientists group. Although 6 out of 11 participants in this group had difficulties understanding the workflow and the results of the workflow, the GUI gave them at least the possibility to try to comprehend the data operations and the feeling to be in control whereas the script-based approach seemed too complex to be comprehended for many. Participants of both groups believed that as non-programmers they would prefer the GUI-tool, as the script-based approach with OpenVibSpec would require too much prior knowledge. Instead, the GUI has used familiar interaction techniques, for example, using drag-and-drop or context menus.
With regard to the exploratory behavior of the participants, no real differences were found between both groups. However, in the life science group there were three participants who performed the same k-Means clustering three times in OpenVibSpec, while in the computer science group there was only one participant who repeated the clustering with the same cluster size three times. All other participants took a more exploratory approach to the task and chose three different cluster sizes (Table 2). In addition, there were also two participants in the life science group who chose a clustering of c=1, which ultimately resulted in a monochromatic image, suggesting that they were not entirely sure what the clustering was doing, at least beforehand. In Orange, no participant attempted exploratory testing of cluster sizes. Instead, the k-means widget was used without modifying the settings, resulting in every participant using the same cluster size (c=5). To get to the settings for the widget, one would have had to open it. However, this was neither requested nor suggested in the task description for Orange. Participants would probably have noticed the cluster size if it had been visually reflected in the widget and perhaps could have been changed directly in the GUI without having to open the settings. Such visual feedback directly visible workflow could promote exploration of different clusterings. However, due to the two-step procedure (first open the widget settings, then change the cluster size), the participants neither knew nor questioned the number of clusters used in the workflow in Orange.
| Group | Orange | OpenVibSpec | ||
|---|---|---|---|---|
| runs | c | runs | c | |
| Life Science | 1 | 5 | 2.3 | 4.4 |
| Computer Science | 1 | 5 | 3 | 4.5 |
Another difference between the groups was that some participants in the life science group were negative about the script-based approach. During task processing with OpenVibSpec, there were at least two participants who clearly verbalized their frustration with the tool, especially when errors occurred. Another participant expressed a strong preference not to have to go through the workflow, but expected the tool to simply analyze the image data and provide him with the results without having to start multiple clusterings or algorithms, so that he would hardly have to interact with the tool at all.
Regarding the perceived efficiency, six participants (four life scientists and two computer scientists) rated Orange as more efficient than OpenVibSpec and two participants (one of each group) rated the efficiency of both tools as equal, although the computing times of Orange were noticeably longer. One possible explanation for this perception is that in OpenVibSpec three runs for k-means were prepared and the majority of participants explored differences between cluster sizes (see Table 2). In addition, the calculations lasted longer when participants used a higher cluster size. Another possible explanation for the perceived efficiency is, that participants (especially from the life science group) felt more comfortable using the graphical widgets.
As planned, the study revealed many usability problems in either of the tools and on different levels of severity. Several issues were discovered as a result of inviting two different user groups in the study. For example, the high barrier entry for non-programmers and script-based workflow management system was particularly evident for the life science group. Here we could show that even the simplest changes in the scripts were challenging for novice users and could only be solved with assistance of the study facilitator. Accordingly, training or tutorials would be necessary for users if they are to apply such script-based tools for their purposes. In addition, some usability problems (including the long loading times for Orange) could already be noticed during the preparation of the study. Ideally, these problems should have been fixed prior to conducting the study, so that the data import could have been performed in both tools.
Another challenge that frequently occurred was that participants had difficulty understanding or interpreting the results. The problem of interpretability could not be traced back exclusively to one principle in the approach selected, but it is particularly likely that either more feedback needs to be worked out based on the methods used. However, it is also possible that the question of bioimaging needs to be more linked to the workflow used rather than the tool.
Considering the amount of effort for the study and the usability problems found, it seems likely that a smaller number of participants would have been sufficient for the results. This illustrates the problem of the number of participants to be used quite well (see Section 0). Probably, even with small-step user tests, severe usability problems would have already been noticed. In part, we have already encountered this during the development of the tasks for the study (e.g., efficiency problems). However, such studies reveal numerous usability issues and requirements for changes, as well as many views and ideas to be interpreted, which, if implemented well, can ultimately lead to better tools and larger user bases.
In order to clarify the importance and benefit of usability tests in the field of WMSs, this study’s methodical approach requires reflection and discussion. One challenge was to design a task that covered roughly the same aspects in two different tools (GUI vs. script). To achieve this,several more modules/plugins had to be integrated into Orange that were not included in the main application. This in turn, only became apparent with the need to create comparative tasks in both tools. The development of the tasks for evaluation therefore already revealed certain issues (e.g. missing modules, efficiency problems). Such usability issues could probably have been found systematically for both tools with a heuristic analysis before the study. Thus, depending on the scope of the analysis and available resources, even a heuristic inspection of the tools could find many problems and opportunities for improvement before conducting a larger scaled user study.
Due to the participation of novice users and the exploratory nature of the study, the results are limited with respect to the use of bioimage processing. Here, in comparison, it would be interesting to see how experienced users working in the field would handle the two tools. In particular, an exploratory strategy, including various clusterings, and the interpretation of results could vary with experienced users. However, a positive aspect of our approach was that different user groups were selected for the study, as both groups highlighted different requirements for the tools.
From a methodological perspective, the formative design of the study was quite appropriate because the tools take very different approaches and the participants had relatively little prior experience in the subject matter. However, it would have been useful to give all participants a brief introduction to Python and Jupyter notebooks beforehand, so that handling the notebooks would not have interfered with the actual execution of the workflow in OpenVibSpec. Additionally, instructions on how to perform different k-means clusterings in Orange would have been helpful, since none of the participant had tried this on their own. On the other hand, multiple clusterings would have further increased the processing time in Orange.
The study benefited from the fact that not only one tool was tested by participants, but that participants used two different tools (GUI vs. script-based) and thus also compared advantages and disadvantages of both approaches to implement workflows. Furthermore, clear preferences between user groups could be identified and partial needs could be revealed. For example, the need to train potential users with programming knowledge became clear, although the task in the study could also be solved without programming knowledge.
Another aspect that could not be considered in this study are learning effects and memorability. Thus, it would be interesting to see how the participants would handle the applications after several weeks, how well they would solve the tasks, and whether the same usability problems would occur again. In addition, one could examine how much learning effort participants without programming expertise would have to put in before they would be able to independently recreate workflows with OpenVibSpec and Orange, and how the perception towards the two tools would change.
This study presented findings from a formative usability study that involved two different user groups solving a specific biomedical image analysis task based on two different implementations, one GUI based and one script-based. With the exploratory nature of this study, specific points of improvements for the different workflow implementations regarding different user groups could be identified. One example for such an improvement is the explanation of workflow results. Whereas research directions such as Explainable AI32 focus on making the internal decisions of AI-powered systems more transparent, this study shows that in the field of bioimaging, explaining the output of the systems could be improved by providing useful information to support the interpretation of the results.
A major limitation of this study certainly lies within the limited prior knowledge of the participants. For future studies, more insight will be gained by involving user groups with more prior knowledge. Also, the study setup only allowed basic comparison between the different user groups and the two implementations, again mainly due to the limited prior knowledge of the participants. Extended future studies may overcome this limitation by conducting a community-wide data challenge accompanied by a usability study of selected participating groups. Besides providing access to significant user groups and allowing a more systematic comparison between user groups and tools, such study may also elucidate the inductive bias of different users and tools for workflows which require a human-in-the-loop.
Zenodo: A formative usability study of workflow management systems in label-free digital pathology - Data and Code24
This repository contains the following underlying data:
• Tutorial-Video.mp4 a tutorial video presented to participants at the beginning of the study
• code/openvibspec/code for script based workflow, including a github snapshot of the OpenVibSpec repository
• orange/GUI based workflow
– workflow-instructions-orange.pdf instructions for orange given to participants
– CompleteWorkflowOrange.ows Orange file that contains the full workflow
• data/openvibspec training and test data for use in openvibspec
• data/orange training and test data for use in Orange
Zenodo: A formative usability study of workflow management systems in label-free digital pathology - Questionaires30
This repository contains the following underlying data:
• data.xlsx demographic data of participants, time needed for task, additional notes
• demographics_questionnaire_de.docx demographic questionnaire in German
• demographics_questionnaire_en.docx demographic questionnaire in English
• interview_de.docx interview questions in German
• interview_en.docx interview questions in English
• maxqda_export_de.xlsx MaxQDA Export file with codes and interview statements in German
Data are available under the terms of the Creative Commons Attribution 4.0 International license (CC-BY 4.0).
AM and TH conceived the project. MJ and EF conducted the usability study. APR and JB prepared software and data for the study. MJ, APR, TH and AM analyzed the outcome of the study. MJ, APR, TH and AM wrote and revised the manuscript.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the rationale for, and objectives of, the study clearly described?
Yes
Is the study design appropriate for the research question?
Partly
Are sufficient details of the methods provided to allow replication by others?
No
Are the datasets clearly presented in a useable and accessible format?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Usability assessment.
Is the rationale for, and objectives of, the study clearly described?
Yes
Is the study design appropriate for the research question?
Partly
Are sufficient details of the methods provided to allow replication by others?
Yes
Are the datasets clearly presented in a useable and accessible format?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Reproducible data analysis, bioinformatics. I would like to express though that I am not an expert on user studies at all. Hence, it should be important to also have a reviewer from that domain!
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 15 Feb 22 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)