Keywords
Staphylococcus aureus, MRSA, Milk, Swab’s hand, Public health
This article is included in the Agriculture, Food and Nutrition gateway.
This article is included in the Pathogens gateway.
Staphylococcus aureus, MRSA, Milk, Swab’s hand, Public health
Based on reviewer comments, we have removed some unnecessary information, we have added some information to the isolation and identification results, we have added the research sample calculation formula to the method, and we have added some research pictures. We have improved the discussion section accordingly. We have corrected the minor typos, and grammar and we want to thanks the reviewers for their valuable comments which improved the quality of manuscript.
See the authors' detailed response to the review by F M Yasir Hasib
See the authors' detailed response to the review by Ikechukwu Benjamin Moses
Staphylococcus aureus is a pathogenic bacteria that can cause public health problems, because these bacteria often contaminate products of animal origin, including milk or commonly known as milk-borne disease (MBD).1 This opportunistic bacterial pathogen that can be found in animals and humans. This bacterium can cause various diseases ranging from mild to systemic skin infections such as pneumonia, arthritis, and meningitis.2–4 In previous studies, S. aureus was mostly transmitted to humans through contaminated milk.5 S. aureus is commonly found on the skin and mucosa of livestock, especially dairy cows with subclinical or clinical mastitis, which is a source of contamination in milk.6 If these bacteria are resistant to beta-lactam antibiotics is referred to as methicillin-resistant S. aureus (MRSA).7
It has been noted in earlier investigations that MRSA can result in new health issues for both people and animals.8 The high rate of MRSA contamination in dairy farms due to excessive administration of antibiotics in the treatment of dairy cows and the spread of these bacteria cannot be separated from sanitation management during milking.3 Contamination can happen from milk that is collected from the udder as well as from the hands of farmers during the milking process.9 The Probolinggo Regency, specifically in Krucil District, is one of the largest milk-producing centers in Indonesia.10 Antibiotics have been widely used as treatment in cases of infection in dairy cattle in Probolinggo, especially in cases of mastitis, so contamination by MRSA in dairy farms in Probolinggo11 is possible.
S. aureus evolved into strain MRSA because it received the insertion of a large DNA element between 20-100 kb called staphylococcal cassette chromosome mec (SCC mec), that underlies the change in normal penicillin-binding protein (PBP), namely PBP2 to PBP2a.12 PBP2a is expressed by the gene encoding mecA contained in SCC mec which has a very low affinity for beta-lactams, so that event cultured on media containing high concentrations of beta-lactams, MRSA survives.13 Molecular detection of the mecA gene using polymerase chain reaction (PCR) is often carried out to confirm the presence of MRSA isolates, but cannot be done in all laboratories because of the ability and cost constraints.14 Constraints in the use of PCR can be replaced by examining MRSA using the disk diffusion method with the antibiotics oxacillin and cefoxitin, which is then continued with an examination using oxacillin resistance screening agar base (ORSAB).15
The purpose of this study was to examine the level of MRSA contamination in dairy cow’s milk and farmer’s hand in Probolinggo, Indonesia, as well as to compare phenotypic detection methods using screening with oxacillin and cefoxitine diffusion disks, ORSAB, and confirming genotypes using PCR to detect mecA-coding genes. The sensitivity and specificity of the test show the effectiveness and ease of application of the MRSA detection method.
Milk samples were taken from the udders of female cows who were in lactation period, while the samples of farmer's hand swabs were taken from farmers who were milking. The sample size in this study refers to the formula used by Regasa et al.16 in the study of the milk safety assessment of Staphylococcus aureus as follows:
Note:
n = Sample size
Z = Z value at 95% confidence level (1.96)16
P = Expected prevalence is 4.8%17
d = Desired absolute precision (4%)
Based on these calculations, 109 milk samples was obtained with the selection of dairy cooperatives purposively based on the amount of milk production in an area and the willingness of dairy cooperatives to participate in the study. Meanwhile, the number of farmer hand swab samples was adjusted to the number of dairy cows owned by each farmer in the dairy cooperative area, of which 41 cattle were obtained from 109 cows.
A total of 109 samples of dairy cow’s milk and 41 samples of farmer’s hand swabs were collected at a dairy farm in the Probolinggo region, East Java, Indonesia from July to September 2021. Dairy cow’s milk samples were taken from each cow in the third press as much as 30 ml which was then stored in a 60 ml sample bottle; the farmer’s hand swab samples were taken from each farmer after the milking process using a sterile cotton swab which was then stored on Amies medium.
As much as 1 ml of each milk sample was put into a 20 ml test tube filled with 9 ml of Mannitol Salt Broth (MSB) medium while for hand swab samples, the Amies medium was vortexed until it became liquid and then 1 ml was added into a 20 ml test tube which has been filled with 9 ml of MSB media. The test tube containing MSB which had been mixed with the sample was incubated in an incubator (Isuzu Model 2-2195, Jica) at 37°C for 24 hours. The samples were cultured and purified using Mannitol Salt Agar (MSA) (Oxoid CM0085) and then incubated at 37°C for 24 hours.
Microscopic examination of bacteria was done through Gram staining to visualise Gram-positive bacteria in the form of cocci and clusters.18 The biochemical examination was carried out using a catalase test and a coagulase test. The catalase test was carried out by dripping 3% hydrogen peroxide (H2O2) on bacterial colonies that had been placed on the surface of the glass.19 The coagulase test was carried out by dripping 200 μl of rabbit plasma into a coagulase test tube containing bacterial colonies, which was then incubated at 37°C for 24 hours.20
The test was carried out following the Clinical and Laboratory Standards Institute (CLSI) 2020 guidelines: S. aureus was tested for susceptibility to the antibiotics oxacillin 1 μg and cefoxitin 30 μg (Oxoid) on Muller Hinton Agar (MHA) plates (Oxoid, CM0337). The identified isolates were purified on mannitol salt agar (HiMedia Pvt. Ltd., M118) and incubated at 37°C for 24 hours. Using a sterile cotton swab (AKD 10903610549), standardized isolates (0.5 McFarland standard) were evenly streaked on the surface of the MHA medium (Oxoid, CM0337). The oxacillin (1 μg) and cefoxitin (30 μg) antibiotic disks were placed side by side with a distance of 50 mm on MHA that had been inoculated with isolates, and then incubated at 37°C for 24 hours to measure the inhibition zone.
S. aureus isolates resistant to oxacillin 1 μg and cefoxitin 30 μg (Oxoid) were confirmed by ORSAB (HiMedia M1415) using S. aureus isolates from the MHA media; plus Oxacillin Resistance Selective Supplement (Supplement, HiMedia Pvt. Ltd., FD191).21
All S. aureus isolates that were resistant to cefoxitin 30 μg and positive on ORSAB examination were then subjected to a PCR test to detect the presence of the mecA gene.22 The DNA extraction process was carried out according to the QIAamp DNA Mini Kit protocol (51304 & 51306), where previously the isolates were purified on MSA (HiMedia Pvt. Ltd, M118) and inoculated on MHA (Oxoid, CM0337). The primer used was mecA F: 5′-AAA ATC GAT GGT AAA GGT TGG C-3′ and mecA R: 5′-AGT TCT GCA GTA CCG GAT TTG C-3′.23 The PCR master mix used GoTaq Green Master Mix (Promega, 9PIM712) which is a ready-to-use solution mixture containing Taq DNA polymerase, dNTPs, MgCl2, and a reaction buffer. DNA was amplified using a Thermal Cycler T100 machine (Bio-Rad, 186-1096) for 40 cycles in 25 μl of the reaction mixture with the following steps: denaturation at 94°C for 30 seconds, annealing at 55°C for 30 seconds, and extension at 72°C for 1 min with a final extension at 72°C for 5 min. A total of 10 μl of PCR product were analyzed by 2% agarose gel electrophoresis, and the gel was visualized under ultraviolet light.24 A positive test indicated a PCR product in the 533-base pair (bp) band.
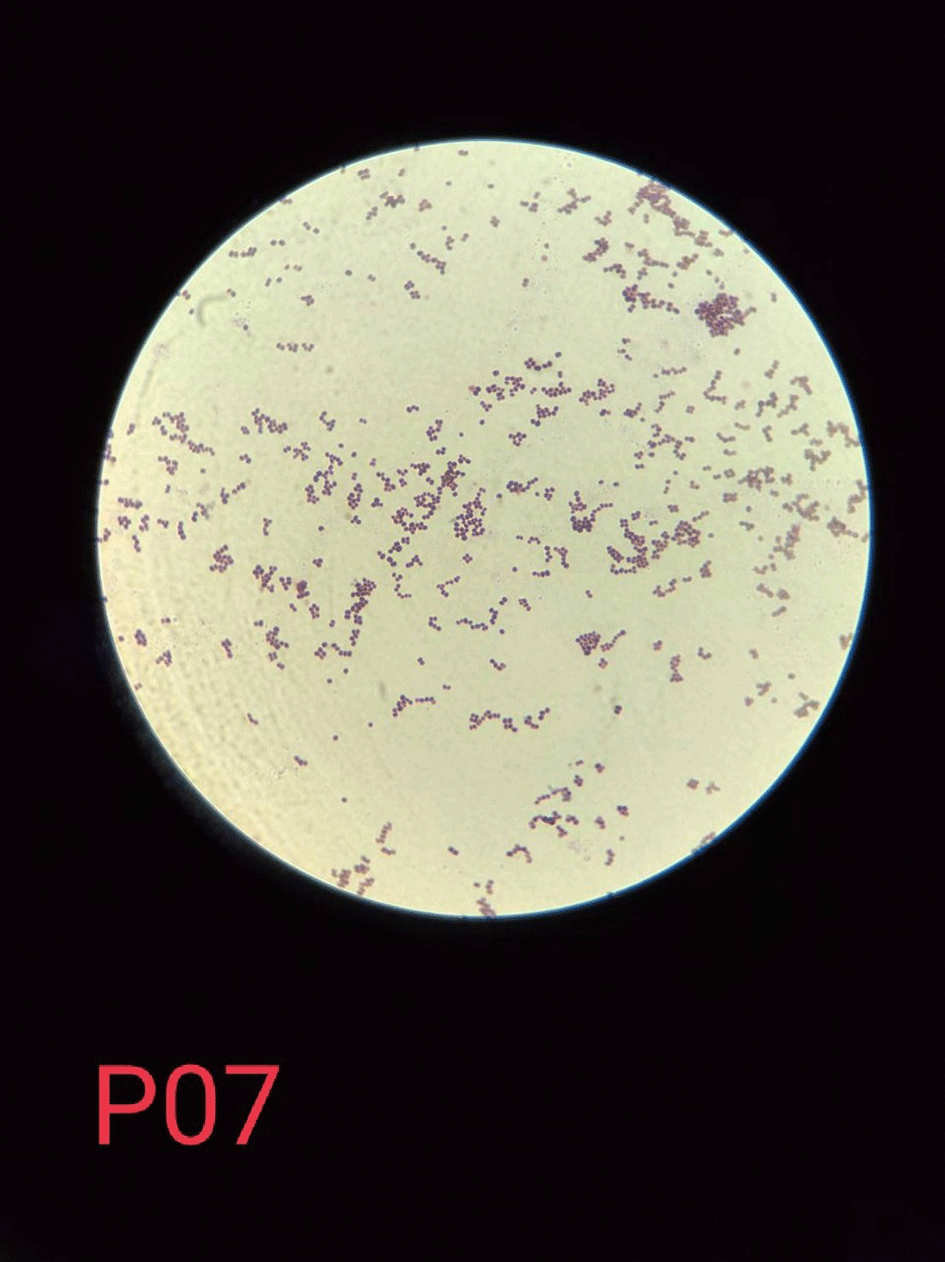
The results of the isolation and identification tests yielded 80 (53.33%) S. aureus isolates from 150 samples taken at a dairy farm in Probolinggo, East Java, Indonesia. The 80 isolates that were positive for S. aureus consisted of 54 isolates from dairy cow’s milk samples and 26 isolates from farmer’s hand swab samples as shown in Table 1. S. aureus had phenotypic colony characteristics on MSA medium, namely a change in color in the medium from red to golden-yellow indicating mannitol fermentation, while the colonies had various pigments including white, golden, and yellow as shown in Figure 1. The Gram staining test showed the Gram-positive colonies in the form of cocci and clusters as shown in Figure 2, which were then confirmed by the catalase test and coagulase test as shown in Figures 3 and 4.19
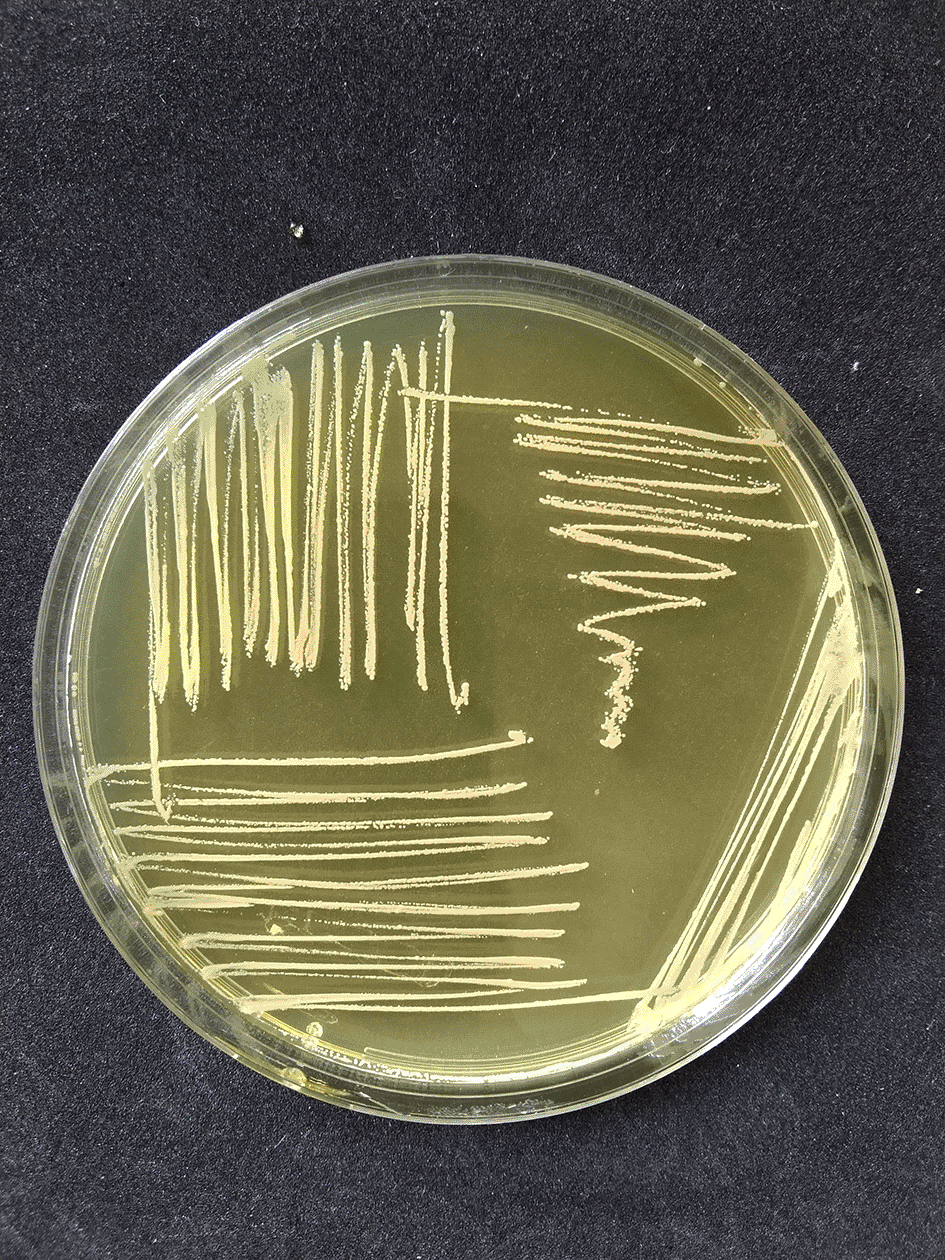
The disk diffusion method on MHA medium showed that 42 isolates exhibited resistance to oxacillin preparations, with a percentage of 52.5% (28 isolates came from dairy cow’s milk samples and 14 isolates came from farmer’s hand swab sample); on the other hand, 10 isolates showed resistance to cefoxitin, with a percentage of 12.5% (five isolates came from dairy cow’s milk samples and five isolates came from farmer’s hand swab samples) as shown in Table 2 and Figure 5.

No S. aureus isolate was found to simply be resistant to cefoxitin, according to the disc diffusion test results, and all isolates that were found to be resistant to cefoxitin were also found to be resistant to oxacillin, as shown in Table 3.
Confirmation of the phenotype test that for resistance to oxacillin and cefoxitin was followed by ORSAB test, with a blue culture coloration indicating positive results while a white coloration indicated negative results. The ORSAB test showed that of the 42 isolates of S. aureus that were resistant to oxacillin, 20 isolates (47.62%) were confirmed MRSA by the disk diffusion method, as shown in Table 4.
| Sample type | Sample code | Number of isolates tested ORSAB (n=42) | Positive ORSAB test |
|---|---|---|---|
| Milk | AS | 28 (66.67%) | 15 (35.71%) |
| Hand swab | AT | 14 (33.33%) | 5 (11.9%) |
| Total | 42 (100%) | 20 (47.62%) |
S. aureus isolates suspected to be MRSA (Phenotypically resistant to cefoxitin and positive for ORSAB) were then tested genotypically using PCR to detect the presence of the gene encoding mecA. A total of 10 isolates suspected to be MRSA were tested, from which three isolates (30% of the total isolates tested by PCR) were detected positive for the mecA gene, as shown in Figure 6. The results of the PCR test showed that isolates suspected to be MRSA were found to have the mecA gene, which is resistant to the antibiotics cefoxitin and oxacillin, as shown in Table 3.
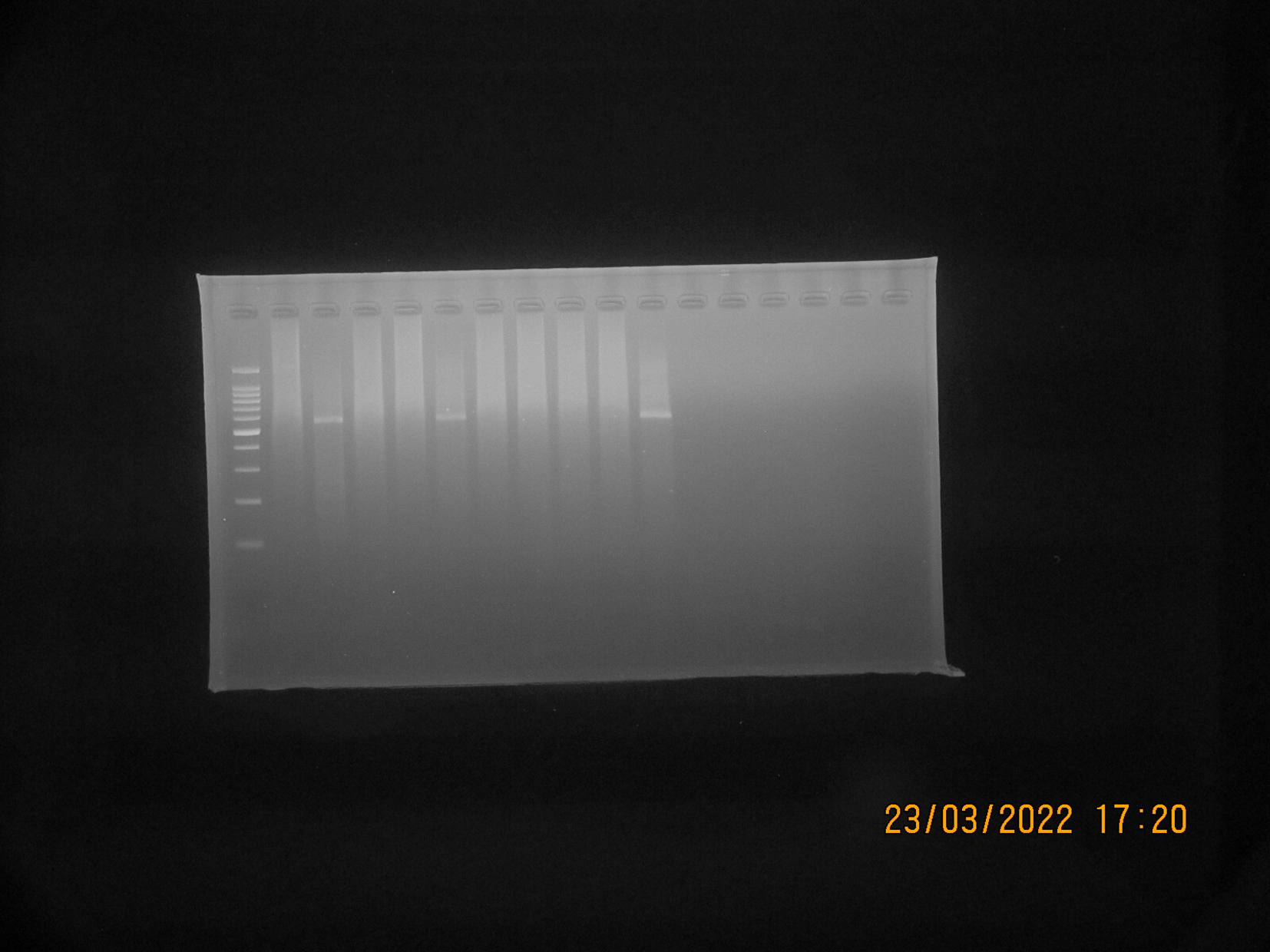
MBD is quite a common public health problem, because it not only has an impact on human health, also has an impact on the health of dairy cows, especially in the milk production and quality sector.25 Several previous studies have reported that the incidence of contaminated milk by S. aureus resistant to antibiotics is found in both developed and developing countries.26 Improper and unhygienic handling of milk, especially during the milking process, plays an important role in the occurrence of milk contamination.27 Unhygienic farmer hands when milking can also potentially transmit pathogenic bacteria in milk, including S. aureus.28
S. aureus is a pathogenic bacterium that can cause various infectious diseases ranging from skin infections to systemic infections that can lead to death.29 In this study, of 150 milk samples, 80 samples (53.33%) were found to have S. aureus contamination; this percentage is higher than the research conducted by Wang et al.30 who isolated 90 (46.15 %) S. aureus from 195 milk samples, and from another study conducted by Jahan et al.31 who isolated 12 (25.53%) S. aureus from 47 milk samples. This study employed a purposive sampling design that was carried out to detect the presence of S. aureus strains in dairy farms that have low milking hygiene, which can increase bacterial contamination in cow’s milk.32 In line with this, the research conducted by Khiabanian et al.33 showed that the difference in the number of isolates found could be influenced by differences in study design such as population and geographic distribution of the sample, infection control practices, and the type of antibiotic used, as seen in Figure 6.
The problem of the incidence of S. aureus infection continues to grow with the emergence of MRSA, which is resistant to all beta-lactam antibiotics, including monobactams and cephalosporins, which are a group of antibiotics often used to treat Staphylococcus infections.34 MRSA infection causes treatment problems and facilitates its spread, so prompt and early diagnosis is needed to identify MRSA accurately.35 In this study, 42 samples (52.5%) of S. aureus were found to be resistant to oxacillin disks, and 10 samples (12.5%) to cefoxitin disks. Miragaia36 stated that the phenotypic detection of MRSA using disk diffusion still has not shown accurate results, and mecA genotyping using PCR is still the main recommendation even though it cannot be done routinely. However, even so, identification of MRSA with disk diffusion is still widely used because it can be done quickly and at a lower cost.37 Diffusion disks using oxacillin and cefoxitin have the same sensitivity level of 100%, and specificities of 74.07% for oxacillin and 92.59% for cefoxitin.38 However, several previous studies reported that the use of the cefoxitin disk diffusion method had a better sensitivity level than that of oxacillin in detecting MRSA, because the oxacillin disk diffusion method still has a high false positive rate.39 Vyas et al.38 stated that false positives could be influenced by beta-lactamase hyperproduction, resulting in the phenotypic expression of oxacillin resistance but without a genotypic resistance mechanism.
In this study, all isolates detected were resistant to the cefoxitin and oxacillin disks. All isolates detected to be resistant to oxacillin and cefoxitin were confirmed by ORSAB assay, in line with a report by Pourmand et al.40 which stated that the ORSAB test has a specificity of 100%. In this study, 20 of the 42 isolates (47.62%) were found to be positive for MRSA. The sensitivity level confirmed the resistance strain being tested while the specificity was to the minimum inhibitory concentration (MIC).41 Cefoxitin-resistant and ORSAB-positive S. aureus isolates were tested genotypically using PCR to detect the presence of the gene encoding mecA; these isolates also had positive results in all phenotypic methods (resistance to cefoxitin and oxacillin in the disk diffusion method and positive results in the ORSAB test). These results are similar to those from research conducted by Ramandinianto et al. 42 The antibiotic cefoxitin is a good inducer for the expression of the mecA gene because it can increase the expression of PBP2a, which is encoded by the mecA gene.43 This also agrees with Reichmann and Pinho44 and Anand et al.45
From this study, it can be concluded that the occurrence of MRSA contamination in milk can be caused by various factors including the unhygienic hands of farmers when milking.46 MRSA contamination poses a serious public health risk, which increases the potential for the spread of difficult-to-treat staphylococci.47 Therefore, microbiology laboratory examinations are very important to isolate and identify MRSA isolates quickly, accurately, and cost-effectively from food samples of animal origin.48 Genotypic detection using PCR to detect the presence of the gene encoding mecA is a molecularly accurate MRSA test; however, in laboratories that cannot perform molecular testing, the cefoxitin disk diffusion method can be used to detect MRSA.49 This is based on the ability of the cefoxitin disk diffusion test in detecting the expression of the mecA gene which can be a more effective and efficient MRSA screening method.50
This study shows that several S. aureus isolates are Methicillin-Resistant S. aureus (MRSA) and have the gene encoding mecA in dairy farms. The spread of S. aureus that is MRSA can be a threat to public health. Thus, prevention and control measures are needed to suppress the spread of S. aureus infection on a dairy farm in Probolinggo, East Java, Indonesia.
Figshare: Detection of mecA gene and methicillin-resistant Staphylococcus aureus (MRSA) isolated from milk and risk factors from the farmer in Probolinggo, Indonesia, https://doi.org/10.6084/m9.figshare.19784005.
This project contains the following underlying data:
Figshare: Detection of mecA gene and methicillin-resistant Staphylococcus aureus (MRSA) isolated from milk and risk factors from the farmer in Probolinggo, Indonesia, https://doi.org/10.6084/m9.figshare.19784005.
This project contains the following extended data:
Data are available under the terms of the Creative Commons Attribution 4.0 International license (CC-BY 4.0).
The authors wish to express their sincere gratitude to Faculty of Veterinary Medicine, Universitas Airlangga. This article was supported in part by the Penelitian Hibah Mandat funding from Universitas Airlangga, Indonesia in the fiscal year 2022, with grant number: 220/UN3.15/PT/2022.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Antibiotic resistance, Public health, Bioinformatics
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Antimicrobial resistance surveillance in veterinary and human medicine, and the development of alternative therapeutic approaches.
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Not applicable
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Antimicrobial resistance surveillance in veterinary and human medicine, and the development of alternative therapeutic approaches.
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
References
1. Clinical and Laboratory Standards Institute: M02 Performance Standards for Antimicrobial Disk Susceptibility Tests, 13th Edition. 2018. 30 Reference SourceCompeting Interests: No competing interests were disclosed.
Reviewer Expertise: Antibiotic resistance, Public health, Bioinformatics
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 3 (revision) 20 Sep 22 |
read | |
|
Version 2 (revision) 12 Sep 22 |
read | |
|
Version 1 30 Jun 22 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)