Keywords
Brain MRI, Canines, ComBat, Data harmonization, Multiple sites, Meningioma, Glioma, Random forest classification
Brain MRI, Canines, ComBat, Data harmonization, Multiple sites, Meningioma, Glioma, Random forest classification
Magnetic resonance imaging (MRI), a powerful technology to detect abnormalities in human and animal organs,1–9 can be challenging for clinically differential diagnosis.10–14 In omics sciences, data normalization (henceforth, “harmonization”) is a crucial preprocessing step prior to downstream analyses,15–21 mitigating any spurious effects on the scientific conclusions incorporated due to undesired sources of variation, such as batch effects, intrinsic factors within the subjects, and scanning sites. Such harmonization is also essential for MRI data, as the signal intensities in these data are measured in arbitrary units that vary across study-visits and patients.22–25
In this study, we demonstrate the effectiveness of a batch-effect correction tool, ComBat,26 widely used in transcriptomics27,28 but also adopted for radiomics data,29,30 in adjusting for undesirable effects of multiple sites on MRI signal intensities (SIs). We chose ComBat due to its superior performance in removing site-specific unwanted variations from fractional anisotropy and mean diffusivity maps in diffusor tensor MRI.29 In their study, the authors considered only controls, used data that were from two “pure” sites, and implemented a sophisticated image-processing pipeline to generate the tissue outcome labels, which resulted in final measurements on the image variables (voxels) having dimensions in the order of 10,000’s. In our case, however, each subject is diseased (meningioma/glioma) and the data come from two “impure” sites, i.e., the “outside” site consists of multiple non-CSU sites, the data thus potentially being noisy due to heterogeneous MRI scanners/protocols used. Notably, such site-heterogeneity can be commonplace to ensure a sufficient sample size. Additionally, we use only three manually recorded image variables, available for all subjects across the sites. Via the downstream performance of the ensemble machine learning classification tool, random forest31–34 (RF), our study thus aims to demonstrate the utility of ComBat harmonization in a “non-ideal” yet practical scenario.
We use n = 244 subjects (dogs) in our study, belonging to one of the following four subpopulations: 1) glioma, scanned at the Colorado State University – Veterinary Teaching Hospital (CSU-VTH), n = 39; 2) glioma, obtained from a site outside CSU, n = 20; 3) meningioma scanned at the CSU-VTH, n = 106; and 4) meningioma, obtained from a site outside CSU, n = 79. Note that we treat the subjects as coming from only two sites -- CSU and “outside”. However, the “outside” site actually consists of 36 unique sites (Table 1).
DN and XY, the two “processors”, generate the data used in the final analyses. DN scans through the conclusion of each patient’s brain MRI diagnostic report stored in the CSU-VTH Philips IntelliSpace PACS (picture archiving and communication system) Radiology software (henceforth referred to as “PACS”) database, labeling the associated brain tumor-type as either “glioma” or “meningioma” based on the radiologist’s/principal interpreter’s conclusion including terms such as “likely”/“most likely”/“most consistent”, etc. Therefore, these binary tumor-type labels are not based on surgical, histopathological evidence and are used as the outcome variable in the downstream RF classification (see the “Statistical analysis” section). Since we do not have access to the diagnostic reports for the subjects from the “outside” site, we consider instead the corresponding ones from the CSU PACS database that are closest to their original exam dates.
For each patient, we only consider the transverse/axial section, T1-weighted, post-contrast scans (typically labeled as “Trans T1 +C”). The processors scan through all the slices within each patient’s respective DICOM file and select up to three representative slices in which the cancerous lesions are most prominently visible (i.e., highest contrast) by naked eye. Note that, among the 244 subjects, we settle with only one suitable slice for seven subjects and two for six subjects (Extended data: Table S1).47 Then, within each chosen slice, two circular regions of interest (ROIs) are drawn encompassing the densest parts visually examined, one each on the lesion and on the “normal” tissue, using the PACS software in-built “drawing” tool. Also note that, as “normal” tissue, we choose facial muscle for seven meningioma subjects and muscle of mastication for the rest (Extended data: Table S2).48 From each of these two ROIs, three statistics for the SIs are noted: mean, standard deviation, and the central point-value. See Figure 1 for an example.
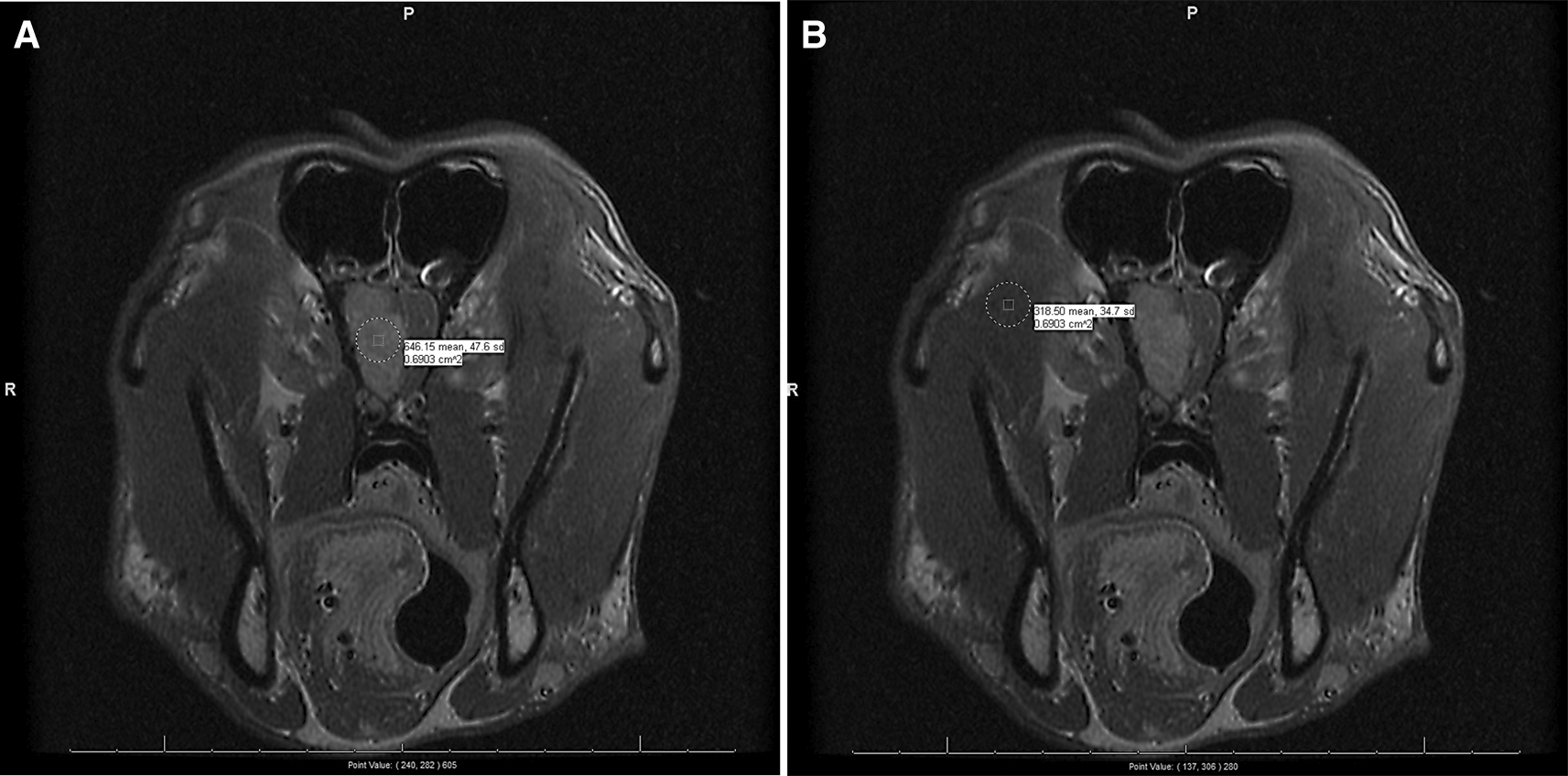
This subject (dog) belongs to the “meningioma outside” subpopulation, i.e., its brain MRI is performed at a non-CSU site and diagnosed with meningioma tumor-type. The normal tissue chosen in (B) is muscle of mastication. The means and the standard deviations of the SIs within the two ROIs are indicated beside the circles drawn and the central point-value SIs are indicated at the bottom of the slides, outside of the parentheses.
Besides the three MRI variables, for each patient we also record the following covariates: three clinical – age (in months) at the time of MRI scan, sex (male, female, male castrated, female spade/spayed), and breedtype; six related to MRI scanner – repetition time (TR), echo time (TE), number of excitations (NEX), slice thickness (mm), frequency phase (X x Y), and field-of-view reconstruction (FOV recon; cm); and one technical – processor.
Note that, for the final analysis, we use both sex and breedtype as binary variables: sex (female/male) and breedtype (non-brachycephalic/brachycephalic). Data on frequency phase are used as two independent scanner covariates. Due to the presence of missing data, we eventually omit the “FOV Recon” scanner covariate from the final analysis. Thus, we have three binary covariates – sex, breedtype, and processor, coded as 0/1; the rest are treated as continuous variables. See Table 2 for a summary of all of the final variables used in our analyses.
The data are grouped based on the four subpopulations as indicated in the columns. Apart from the three binary covariates – sex, breedtype, and processor – that are coded as 0/1, the rest are treated as continuous variables; each cell-value indicates the range in the top line and the median (median absolute deviation in parentheses) in the bottom line.
Preprocessing of the data and final variables
For each of up to three selected slices corresponding to each sample, we first normalize the mean, the standard deviation, and the central point-value of the SIs within the diseased ROI by taking respective ratios to the normal ROI within that same slice (Figure 1). We call these three measures adj-mean (SI), adj-SD (SI), and adj-cent (SI), respectively. Next, for each sample, we compute the means of these adjusted measures across the selected slices. These three summarized measures, respectively referred to as μ (adj-mean (SI)), μ (adj-SD (SI)), and μ (adj-cent (SI)), are used as the final three image variables in the subsequent analyses (Figures S1 and S2).50–55 The intercorrelations among the three continuous image variables and the disease labels (0 = glioma, 1 = meningioma) are shown in Figure S3.56,57 We note that, for both CSU and outside sites μ (adj-mean (SI)) and μ (adj-cent (SI)) are maximally correlated with the disease labels and the correlations among the μ (adj-SD (SI)) and disease labels are negligible. Among the continuous covariates across both sites, while age (in months), μ (adj-mean (SI)), and μ (adj-cent (SI)) resemble a Gaussian distribution, those of others deviate greatly from it (data not shown).
For the classification of meningioma and glioma brain-tumors (glioma treated as the “positive” class), we apply RF31–34 and evaluate classification performance based on sensitivity, specificity, and total accuracy, benchmarked via “lower” and “upper” bounds (Table 3). Using the same site for training and test sets, we expect better RF classification performance (upper bound) compared to when using different sites (lower bound).
M: Meningioma, G: Glioma. For “lower” bound computations, we use all the samples within the outside site (n = 99, M/G = 79/20) to train the RF model, and randomly subsample n = 38 subjects from the CSU population, ensuring M/G = 19/19 representation, for the test set. For “upper” bound computations, we randomly subsample n = 79 meningioma CSU subjects from the remaining 87 for the training sets and use the same test sets as those used for the lower bounds.
| Training set (n = 99, M/G = 79/20) | Test set (n = 38, M/G = 19/19) | |
|---|---|---|
| Lower bound | Outside | CSU |
| Upper bound | CSU | CSU |
For the “lower” bound calculations, we use all the samples within the outside site (n = 99, M/G = 79/20) to train the RF classifier, and randomly subsample n = 38 subjects from the CSU population, ensuring M/G = 19/19 representation, for the test set. Note that, the training set for the lower bound have 4:1 imbalanced class distribution in the outcome, which we adjust for using the Synthetic Minority Oversampling TEchnique (SMOTE),35 using arguments perc.over = 3 and perc.under = 1.45 within the smote() function. The size of a final training set is thus increased to n = 159 (M/G = 79/80). We use the original n = 79 meningioma samples and the n = 80 glioma cases that are generated using SMOTE. Within this training set, we tune the parameters of the RF classifier using 5-fold cross-validation repeated 25 times, and using all possible combinations of predictor variables in the model via the mtry argument in the train() function. For the “upper” bound calculations, we keep the identical test set compositions as in lower bound computations, and form the training set by randomly subsampling n = 79 “meningioma CSU” subjects from the remaining 87. We repeat this exercise of computing lower and upper bounds 75 times, each time with a different training-test split. Finally, we report the medians (and median absolute deviations) of the classification metrics across these 75 random samples; see Table 5 for an example.
We investigate the RF classifier performance at the lower and upper bounds for the following scenarios:
• [Case 0: one scenario] We examine the effectiveness of using three clinical covariates only in classifying the tumor types. No image, technical, and scanner covariates are used, and therefore, no ComBat harmonization is involved.
• [Case 1: four scenarios] We use the three image variables in ComBat. Besides, we either use the three clinical covariates or not in ComBat and in subsequent RF, thus giving rise to four scenarios (a – d; Table 4). We do not use any technical and scanner covariates in ComBat.
| ComBat: 3 Clinical covariates | |||
|---|---|---|---|
| No | Yes | ||
| Random Forest: 3 Clinical covariates | No | Scenario a | Scenario b |
| Yes | Scenario d | Scenario c | |
To assess the impact of ComBat harmonization on RF classification performance, we conduct nonparametric tests (Wilcoxon’s signed-rank paired one-sided tests with continuity correction) to examine whether a post-ComBat classification metric lower bound is: (1) significantly greater than that for its pre-ComBat counterpart, and (2) significantly lower than the corresponding upper bound (Table 5). Glioma is treated as the “positive” class in classification and, therefore, sensitivity measures the proportion of true glioma cases correctly identified, specificity measures the proportion of true meningioma cases correctly identified, and total accuracy measures the total proportion of true meningioma and glioma cases correctly identified.
The medians and median absolute deviations of the classifiation metrics are computed based on 75 repetitions of random training/test splits. Values closer to 1 indicate better performance. For post-ComBat lower bounds: 1) bold indicates significantly greater value (p-value < 0.05, Wilcoxon’s signed-rank paired one-sided test with continuity correction) compared to the corresponding pre-ComBat lower bound; 2) underline indicates corresponding upper bound is not significantly higher. Therefore, bold and underline together indicate the best results using ComBat.
Below we discuss the full set of results for the scenarios in Cases 0 and 1.43–46,50–57 Note that, besides these two cases, we also examine the results of another case (Case 2) in which, alongside the three image variables, we include one technical covariate and six scanner covariates (see the “Study population and data generation” section) in the ComBat step. However, since the essence of these results is mostly similar to that of Case 1, we set them aside as “Extended data” (Extended data: Table S3).49
Using only the clinical covariates of the subjects in the RF model (Case 0), the lower bound total accuracies are not significantly lower than those for upper bounds: both medians = 57.9%; p-value = 0.332 (Figure 2). The lower bounds of the sensitivity and the specificity measures are also not significantly lower than those for the upper bounds: p-values 0.133 and 0.884 respectively. Thus, the distributions of the age/sex/breed-type between meningioma/glioma subjects do not vary significantly across sites. For example, exact p-values corresponding to the Pearson’s chi-squared tests (with Yates’ continuity correction) on the two 2×2 contingency tables for sex and breed-type distributions across CSU and Outside sites are 0.762 and 0.604, respectively. Also, among all scenarios, RF achieves the lowest medians of total accuracy and sensitivity in this case, which indicates an overall poor predictive strength of using only clinical covariates in the RF model (Figures 2 and 3).
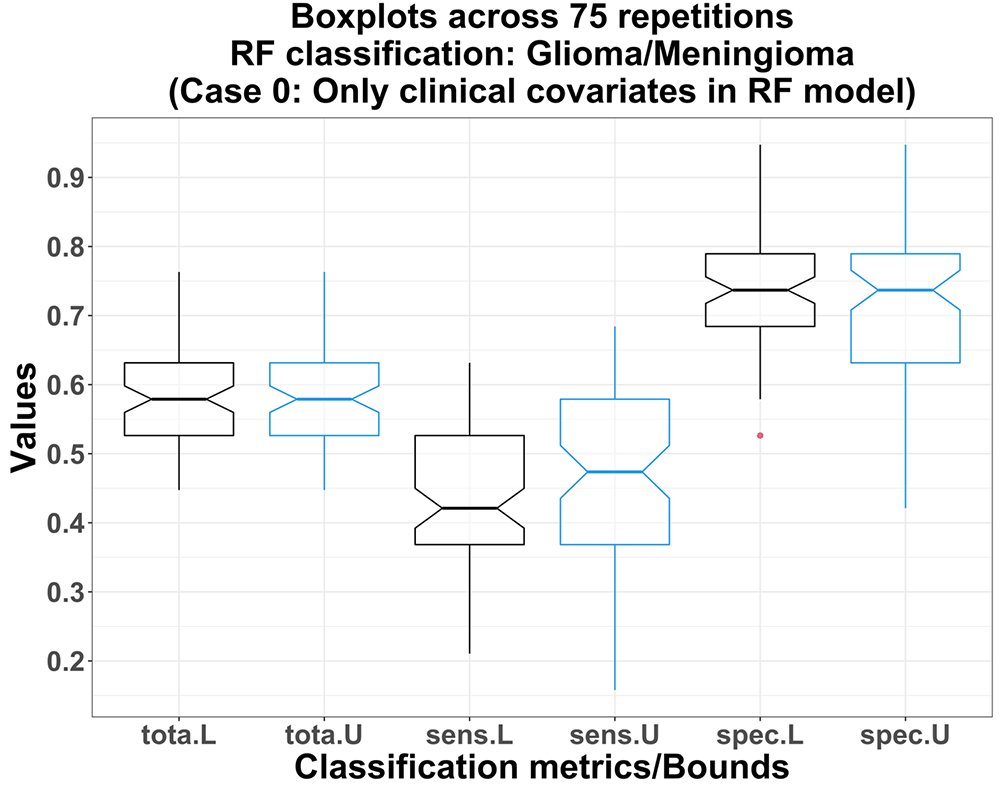
L, U: lower bound (black) and upper bound (blue) obtained from RF models using only three clinical covariates.
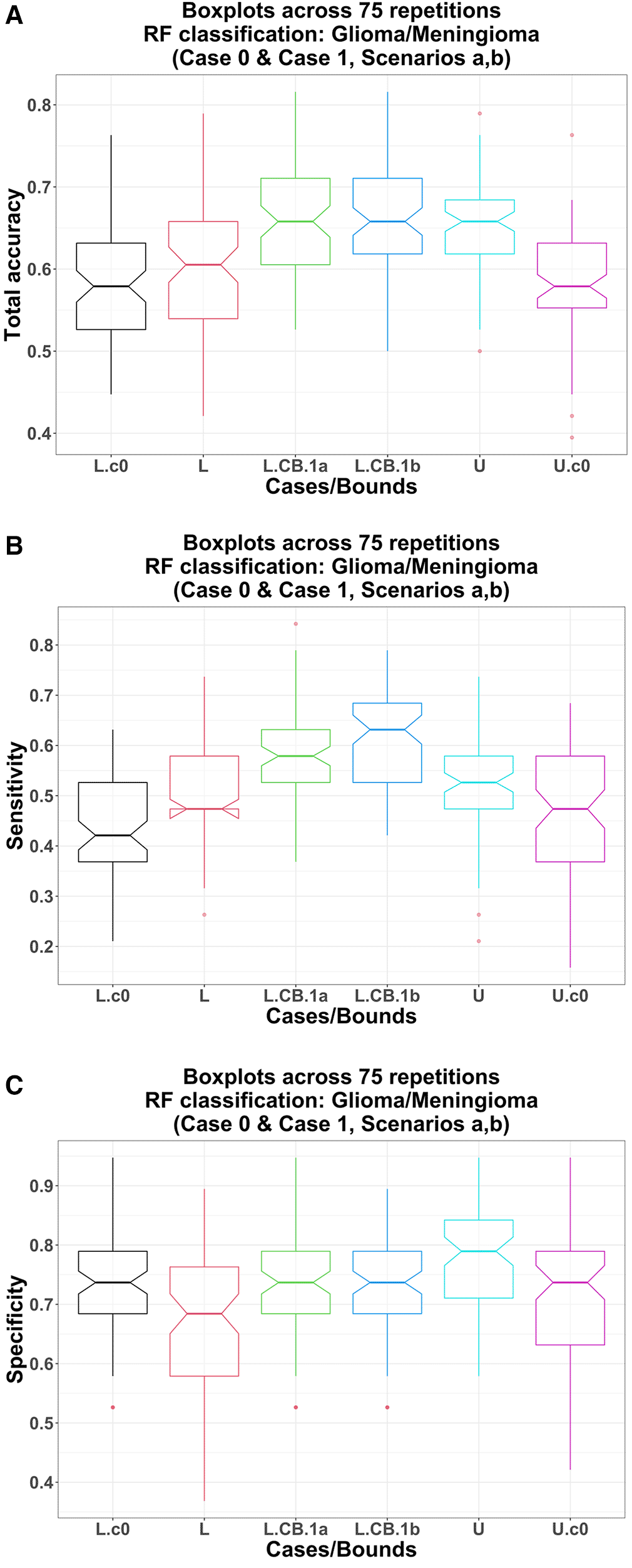
L.c0, U.c0: lower bound (black) and upper bound (magenta) obtained from RF models using only three clinical covariates; no ComBat harmonization involved; L, L.CB, U: pre-ComBat lower bound (red), post-ComBat lower bounds (green, 1a; blue, 1b), and upper bound (cyan) obtained from RF models using only three image variables.
Pre-harmonization
Total accuracy: Using only the image variables in the RF model, the lower bound total accuracy (pre-ComBat) does not differ significantly from that using only three clinical covariates (Case 0): medians 60.5% vs. 57.9%; p-value = 0.270. However, the upper bound total accuracy is significantly higher than that in Case 0: medians 65.8% vs. 57.9%; p-value = 4.06 E-07 (Figure 3-A).
Sensitivity: Using only the image variables in the RF model, the lower bound sensitivity (pre-ComBat) is significantly higher than that using only three clinical covariates (Case 0): medians 47.4% vs. 42.1%; p-value = 9.68 E-04. Similarly, the upper bound sensitivity is also significantly higher than that in Case 0: medians 52.6% vs. 47.4%; p-value = 6.58 E-04 (Figure 3-B).
Specificity: Using only the image variables in the RF model, interestingly, the lower bound specificity (pre-ComBat) is significantly lower than that using only three clinical covariates (Case 0): medians 68.4% vs. 73.7%; p-value = 3.31 E-03. However, the upper bound specificity is significantly higher than that in Case 0: medians 78.9% vs. 73.7%; p-value = 5.67 E-05 (Figure 3-C).
Post-harmonization
Total accuracy: Using post-ComBat harmonization (scenarios a, b), the total accuracy lower bounds are significantly higher compared to their pre-ComBat and Case 0 counterparts. For example, post-ComBat with only three image variables (scenario a): (1) vs. pre-ComBat: medians 65.8% vs. 60.5%; p-value = 2.64 E-08 (Table 5, Figure 3-A) and (2) vs. using only the clinical covariates (Case 0): medians 65.8% vs. 57.9%; p-value = 4.98 E-08 (Figure 3-A).
Sensitivity: Using post-ComBat harmonization (scenarios a, b), the sensitivity lower bounds are significantly higher compared to their pre-ComBat and Case 0 counterparts. For example, post-ComBat with only three image variables (scenario a): (1) vs. pre-ComBat: medians 57.9% vs. 47.4%; p-value = 4.33 E-08 (Table 5 and Figure 3-B) and (2) vs. using only the clinical covariates (Case 0): medians 57.9% vs. 42.1%; p-value = 7.88 E-11 (Figure 3-B).
Specificity: Using post-ComBat harmonization (scenarios a, b), the specificity lower bounds are significantly higher compared to their pre-ComBat counterparts. For example, post-ComBat with only three image variables (scenario a) vs. pre-ComBat: medians 73.7% vs. 68.4%; p-value = 1.16 E-03 (Table 5 and Figure 3-C). Interestingly though, these post-ComBat lower bounds are not significantly higher than that using only the clinical covariates (Case 0): all three medians 73.7%; p-values (scenarios a and b vs. Case 0) = 0.347 and 0.359, respectively (Figure 3-C).
These results confirm that using just the three image variables in the RF model, ComBat harmonization enhances the RF classification performance (except for specificity) compared to that in pre-ComBat and when using only the clinical covariates.
Pre-harmonization
Total accuracy: Using the image variables and the clinical covariates in the RF model, the lower bound total accuracy (pre-ComBat) is significantly higher than that using only three image variables in RF: medians 68.4% vs. 60.5%; p-value = 7.48 E-09. Similarly, the upper bound total accuracy is also significantly higher: medians 71.1% vs. 65.8%; p-value = 3.64 E-07 (Table 5, Figure 4-A).
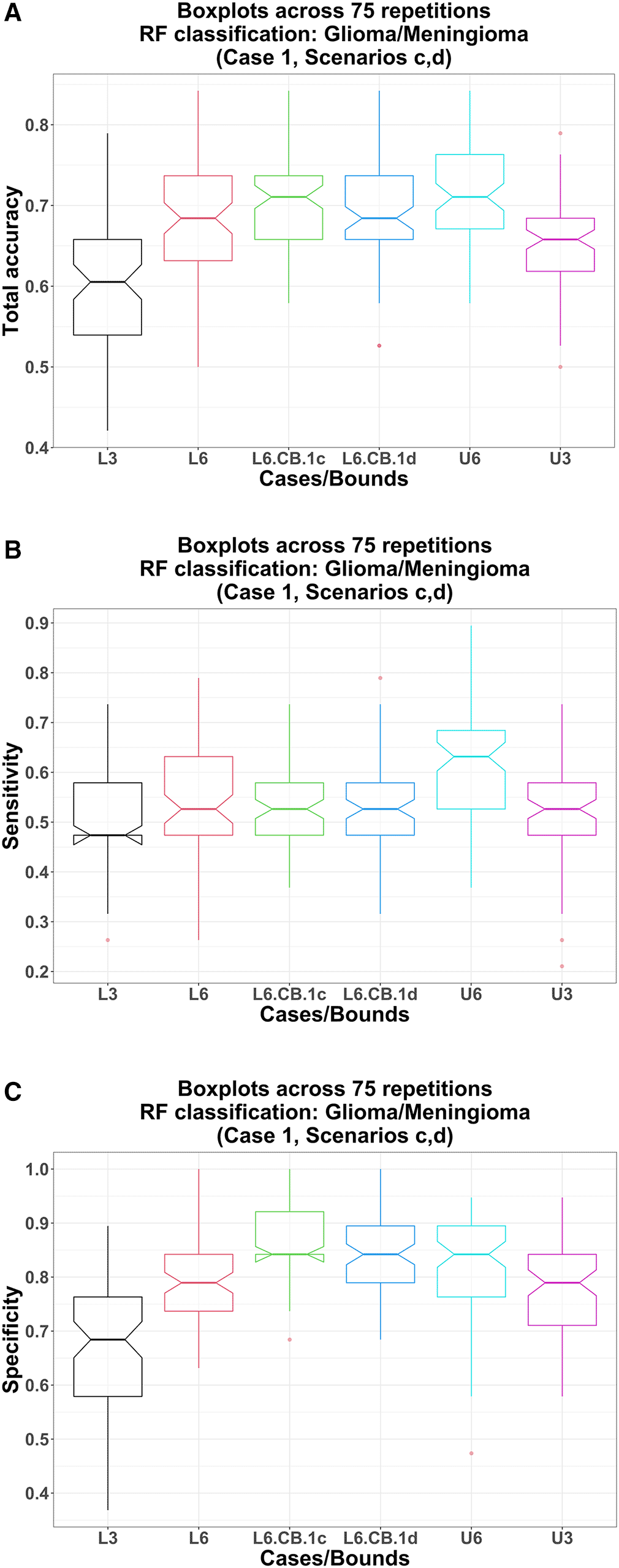
L3, U3: pre-ComBat lower bound (black) and upper bound (magenta) obtained from RF models using only three image variables; L6, L6.CB, U6: pre-ComBat lower bound (red), post-ComBat lower bounds (green, 1c; blue, 1d), and upper bound (cyan) obtained from RF models using three image variables and three clinical covariates.
Sensitivity: Using the image variables and the clinical covariates in the RF model, the lower bound sensitivity (pre-ComBat) is significantly higher than that using only three image variables in RF: medians 52.6% vs. 47.4%; p-value = 8.77 E-04. Similarly, the upper bound sensitivity is also significantly higher: medians 63.2% vs. 52.6%; p-value = 1.76 E-06 (Table 5, Figure 4-B).
Specificity: Using the image variables and the clinical covariates in the RF model, the lower bound specificity (pre-ComBat) is significantly higher than that using only three image variables in RF: medians 78.9% vs. 68.4%; p-value = 2.33 E-10. Similarly, the upper bound specificity is also significantly higher: medians 84.2% vs. 78.9%; p-value = 2.90 E-03 (Table 5, Figure 4-C).
Post-harmonization
Total accuracy: Using post-ComBat harmonization (scenarios c, d), the total accuracy lower bounds are significantly higher compared to their pre-ComBat and post-ComBat with only image variables in RF counterparts. For example, post-ComBat using three image variables and three clinical covariates (scenario c): (1) vs. pre-ComBat: medians 71.1% vs 68.4%; p-value = 8.80 E-04 and (2) vs. using only image variables in the RF model (scenario b): medians 71.1% vs. 65.8%; p-value = 1.84 E-04. Moreover, comparing between post-ComBat scenarios c and d: medians 71.1% vs 68.4%, p-value = 6.97 E-03 (Table 5, Figure 4-A).
Sensitivity: Using post-ComBat harmonization (scenarios c, d), the sensitivity lower bounds are not significantly higher compared to their pre-ComBat counterparts. For example, post-ComBat using three image variables and three clinical covariates (scenario c) vs. pre-ComBat: both medians 52.6%; p-value = 0.953 (Table 5, Figure 4-B). However, this post-ComBat sensitivity lower bound in scenario c is significantly higher than that using only image variables (scenario d): both medians 52.6%; p-value = 0.0177. Interestingly, post-ComBat sensitivity in scenario c (and d) deteriorates significantly compared to those when not using the clinical covariates in the RF model in scenario b (and scenario a): medians 52.6% vs. 63.2% (52.6% vs. 57.9%); p-value = 2.07 E-05 (6.93 E-05; Table 5).
Specificity: Using post-ComBat harmonization (scenarios c, d), the specificity lower bounds are again significantly higher compared to their pre-ComBat counterparts. For example, post-ComBat specificity lower bound using three image variables and three clinical covariates (scenario c) vs. pre-ComBat: medians 84.2% vs. 78.9%; p-value = 9.44 E-10 (Table 5, Figure 4-C). This post-ComBat specificity lower bound in scenario c is also significantly higher than that using only image variables (scenario d): both medians 84.2%; p-value = 2.69 E-03 (Table 5, Figure 4-C) and compared to those when not using the clinical covariates in the RF model (scenario b): medians 84.2% vs. 73.7%; p-value = 3.05 E-12 (Table 5).
These results confirm that using the image variables and clinical covariates together in the RF model, with or without ComBat harmonization, results in better RF classification performance (except for sensitivity) than using only the image variables. Furthermore, using the image variables as well as the clinical covariates in both ComBat harmonization and the RF model provides the highest total accuracy and specificity across all scenarios.
In this case-study, we demonstrate the efficacy of MRI data harmonization using ComBat in enhancing the downstream RF classification performance. Utilizing the clinical covariates along with the image variables both in ComBat and RF (Case 1, scenario c) results in the highest total accuracy. When adjusting for the technical and scanner covariates in ComBat (Case 2), we only notice significant improvements in specificity (correct identification of true meningioma cases; scenarios c, d) compared to when not using them (Case 1; Tables 5 and S3). For both cases, RF achieves the highest specificity with the clinical covariates included in the model, irrespective of including them in ComBat (e.g., maximum median value for Case 1 is 84.2%, scenarios c, d; Table 5). Of all cases and scenarios, RF attains the highest sensitivity (correct identification of true glioma cases) when we include the clinical covariates in ComBat but not in the classification model in Case 1 (maximum median value is 63.2%, scenario b; Table 5).
In summary, we confirm the overall effectiveness of ComBat harmonization in adjusting for the site-specific variability even for our “non-ideal” as a practically feasible, noisy, low-dimensional, manually processed MRI dataset.
The highest median total accuracy we obtain is 71.1% (Case 1, scenario c). However, among the 75 repetitions, we do notice up to a maximum of 84.2%. The challenge in attaining any higher total accuracy is mainly poised by low sensitivity, i.e., correct identification of true glioma cases, possibly due to: 1) insufficient predictors – we have used three available, manually generated image variables and three covariates for our analyses; 2) the possible minor mislabeling of the tumor-types or imprecise ROIs because the labels are based on the visual inspection and subjective, expert conclusion of the examining radiologists at the CSU-VTH and not confirmed via surgical histopathology, or because the ROIs in each scan-slice are drawn by two non-radiologists, and hence can possibly incur imprecise diseased/normal ROIs; 3) non-homogeneous sites – ComBat performance can potentially sharpen further with more homogeneous composition of the “outside” site; 4) an imbalanced outcome classes – although we address the severe class imbalance, a more balanced distribution in the original data may enhance RF performance36; and 5) the choice of class imbalance adjustor and classifier – one may choose a different class-imbalance adjustment, such as “over-sampling”,37 or a different classifier, such as logistic regression.38 However, our initial exploration suggests that the SMOTE-RF combination provides better results than those of some other alternatives (data not shown).
Figshare: Image and Covariates Data on CSU-Meningioma Subjects. https://doi.org/10.6084/m9.figshare.19497671.v1.43
Figshare: Image and Covariates Data on CSU-Glioma Subjects. https://doi.org/10.6084/m9.figshare.19497683.v1.44
Figshare: Image and Covariates Data on Outside-Meningioma Subjects. https://doi.org/10.6084/m9.figshare.19497686.v1.45
Figshare: Image and Covariates Data on Outside-Glioma Subjects. https://doi.org/10.6084/m9.figshare.19497692.v1.46
Figshare: Table S1: Number of Subjects with Less Than Three Image Slices Selected. https://doi.org/10.6084/m9.figshare.19497701.v3.47
Figshare: Table S2: Number of Subjects for Whom Facial Muscle is Used as Normal Tissue. https://doi.org/10.6084/m9.figshare.19497707.v2.48
Figshare: Table S3: Case 2 Full Results. https://doi.org/10.6084/m9.figshare.19498832.49
Figshare: Figure S1-A. https://doi.org/10.6084/m9.figshare.19498934.v1.50
This project contains the following extended data:
• New_CSUOut-MeninGlio_boxplot_final_meanSI.png (Boxplots of means [across up to three slices] of normalized mean of signal intensities measured on 244 subjects distributed across four subpopulations).
Figshare: Figure S1-B. https://doi.org/10.6084/m9.figshare.19498937.v1.51
This project contains the following extended data:
• New_CSUOut-MeninGlio_boxplot_final_sdSI.png (Boxplots of means [across up to three slices] of normalized standard deviation of signal intensities measured on 244 subjects distributed across four subpopulations).
Figshare: Figure S1-C. https://doi.org/10.6084/m9.figshare.19498940.v1.52
This project contains the following extended data:
• New_CSUOut-MeninGlio_boxplot_final_centSI.png (Boxplots of means [across up to three slices] of normalized central point-value of signal intensities measured on 244 subjects distributed across four subpopulations).
Figshare: Figure S2-A. https://doi.org/10.6084/m9.figshare.19498943.v1.53
This project contains the following extended data:
• Processors_allGroups_boxplot_final_meanSI.png (Boxplots of means [across up to three slices] of normalized mean of signal intensities measured by two processors [“XY” and “DN”] on 244 subjects distributed across four subpopulations: GC = “Glio-CSU”, MC = “Menin-CSU”, GO = “Glio-Out”, MO = “Menin-Out”).
Figshare: Figure S2-B. https://doi.org/10.6084/m9.figshare.19498946.v1.54
This project contains the following extended data:
• Processors_allGroups_boxplot_final_sdSI.png (Boxplots of means [across up to three slices] of normalized standard deviation of signal intensities measured by two processors [“XY” and “DN”] on 244 subjects distributed across four subpopulations: GC = “Glio-CSU”, MC = “Menin-CSU”, GO = “Glio-Out”, MO = “Menin-Out”).
Figshare: Figure S2-C. https://doi.org/10.6084/m9.figshare.19498949.v1.55
This project contains the following extended data:
• Processors_allGroups_boxplot_final_centSI.png (Boxplots of means [across up to three slices] of normalized central point-value of signal intensities measured by two processors [“XY” and “DN”] on 244 subjects distributed across four subpopulations: GC = “Glio-CSU”, MC = “Menin-CSU”, GO = “Glio-Out”, MO = “Menin-Out”).
Figshare: Figure S3-A. https://doi.org/10.6084/m9.figshare.19498952.v1.56
This project contains the following extended data:
• meninglioCSU_corr_final_3img_dislab.png (Pearson’s correlations among the three image variables and the disease labels [“dis.lab”; meningioma = 1, glioma = 0] within CSU subjects).
Figshare: Figure S3-B. https://doi.org/10.6084/m9.figshare.19498964.v1.57
This project contains the following extended data:
• meninglioOut_corr_final_3img_dislab.png (Pearson’s correlations among the three image variables and the disease labels [“dis.lab”; meningioma = 1, glioma = 0] within “Outside” subjects).
Data are available under the terms of the Creative Commons Zero “No rights reserved” data waiver (CC0 1.0 Public domain dedication).
Source code available from: https://github.com/KechrisLab/ComBat_dogBrainMRI/tree/MRI
Archived source code at time of publication: https://doi.org/10.5281/zenodo.6632525.58
License: GNU General Public License v3.0
We generated all imaging data using the Philips IntelliSpace PACS Radiology software v4.4 (Philips Healthcare Informatics, Inc, 4100 East Third Avenue, Suite 101, Foster City, CA 94404, USA); license purchased by the CSU-VTH. We performed all of the statistical analyses and generate all of the figures using the R statistical software, version 4.1.0.39 We implemented the ComBat data harmonization using the neuroCombat R software package, which is publicly available in Jean-Philippe Fortin’s GitHub: https://bit.ly/fortin-ComBat-git, and the SMOTE imbalanced class adjustment using the smote() function within the performanceEstimation CRAN package.40 For the RF classifier, we use method = “rf” input argument in the train() function and compute the classification performance evaluation metrics using the confusionMatrix() function, both within the caret CRAN package.41,42 As a freely available alternative to PACS for a DICOM viewer and imaging data generator, we suggest Horos.
Approval of VCS #2018-162 “Lymphotropic Nanoparticle Enhanced MRI for Diagnosis of Metastatic Disease in Canine Head and Neck Tumors” was obtained by Dr. Lynn Griffin on June 4, 2018, and subsequently on August 8, 2019 (for amendment to increase the approved animal numbers), from the Colorado State University Veterinary Teaching Hospital Clinical Review Board. The Clinical Review Board consists of 14 faculty members (as of August 8, 2019) from the College of Veterinary Medicine and Biomedical Sciences including a standing member of IACUC, the Hospital Director, and the Chair of the Department of Clinical Sciences.
Client consent was obtained from the respective owners of all dogs included in this study to use all obtained images and medical data for the purposes of research. Consent for publication is not applicable.
We are grateful to Kevin Kirsch of Colorado State University for providing us generous support on the setup of and access to CSU remote workstation and the Philips IntelliSpace PACS software and promptly clarifying our doubts as and when they appeared. We also thank Debashis Ghosh of Colorado School of Public Health and Natalie Serkova of University of Colorado School of Medicine for providing valuable suggestions and recommendations.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the background of the case’s history and progression described in sufficient detail?
Partly
Is the work clearly and accurately presented and does it cite the current literature?
Partly
If applicable, is the statistical analysis and its interpretation appropriate?
I cannot comment. A qualified statistician is required.
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Is the case presented with sufficient detail to be useful for teaching or other practitioners?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: This article covers methods and data analysis that are outwith my area of expertise (which is in veterinary clinical neurology and basic cellular neuroscience).
Is the background of the case’s history and progression described in sufficient detail?
Partly
Is the work clearly and accurately presented and does it cite the current literature?
Partly
If applicable, is the statistical analysis and its interpretation appropriate?
I cannot comment. A qualified statistician is required.
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Partly
Is the case presented with sufficient detail to be useful for teaching or other practitioners?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Veterinary clinical research; bench research in neuroscience
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 07 Jul 22 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)