Keywords
gene polymorphism, renal transplant rejection, cytotoxic T-lymphocyte associated protein 4 (CTLA-4)
This article is included in the Cell & Molecular Biology gateway.
gene polymorphism, renal transplant rejection, cytotoxic T-lymphocyte associated protein 4 (CTLA-4)
High mortality and morbidity rates are still the main issues in end-stage renal disease (ESRD), and it increases healthcare utilization.1 There are several options directed for renal replacement therapy as ESRD treatment, which are hemodialysis (HD), continuous-ambulatory peritoneal-dialysis (CAPD), and renal transplantation. For now, renal transplantation is the gold standard treatment for ESRD.1,2 Renal transplantation is still the best method option for treatment for ESRD due to better quality of life, the modifiable morbidity rate, promising survival rates, and the greatest impacts in daily basis activities in spite of the rejection risks as one of the complications.3,4
Presently, renal transplantation is the most effective treatment for end-stage renal illness, because it preserved individuals with ESRD’s lives.2 However, the big issue of renal transplantation is acute rejection in some recipient cases, which diminishes the quality of the donated kidney.2 Renal transplantation rejection can consequently decrease the renal physiological function and become a threat to a patient’s life. Therefore, it is essential to investigate factors that aggravate transplantation rejection.
Cytotoxic T-lymphocyte antigen-4 (CTLA-4), is a regulatory molecule that inhibits T-cell effector action after first costimulatory signal activation.3,4 CTLA-4 has been implicated in acute renal transplantation rejection etiology, according to available dataset.3 The CTLA-4 +49A/G single nucleotide polymorphism (SNPs) involves a shift from the A to the G-allele that leads to several complicated sequences that jeopardize the next transcription-translation phase of DNA. CTLA-4 is a key CTLA variation, and current findings indicate that the CTLA-4 +49A/G SNPs increases the likelihood of rejection. Previous meta-analysis has been conducted to evaluate the association between the CTLA-4 +49A/G SNPs and the incidence of renal transplantation rejection.4 Nonetheless, there was no assessment of the nature of polymorphism, which included Hardy-Weinberg equilibrium in order to exclude the chance of polymorphism influenced by evolution, moreover only limited studies were included. This study was conducted to determine if the CTLA-4 +49A/G SNPs were connected with the incidence of renal transplantation rejection by compiling a large number of previously published papers.
The study was reported based on the Preferred Reporting Items for Systematic Reviews and Meta-analysis (PRISMA) guidelines.5 The search was conducted on February 2nd, 2022. Retrospective and prospective analytical randomized control trials (RCTs) published prints from Embase, PubMed, Cochrane, and Web of science were included in accordance with the PRISMA guideline and thoroughly analyzed using a random or fixed effect model regarding to its heterogeneity. These datasets were populated with the retrieval approach “(cytotoxic-T-lymphocyte-antigen-4 OR CTLA-4) AND (gene polymorphism OR single nucleotide polymorphisms OR allele OR alleles OR genotype OR genotypes) AND (renal OR kidney) AND (transplant OR transplantation) AND (acute rejection).” The extra reports were discovered using the citations found in the selected papers.
Studies included had quantitative data of renal transplantation rejection genotype that had the cases of acute rejection and non-acute rejection comparison data; and adequate data on the distribution of the CTLA-4 +49A/G genotype was required. We also include retrospective or prospective studies, randomized trials, that provided sufficient data of odds ratio (OR) 95% confidence interval (CI). The thirteen studies that were included are shown in Figure 1. Other studies than those stated before were excluded.
We also measured the Hardy-Weinberg Equilibrium (HWE) formula (X2>3.84 indicated as deviation from HWE) as described by Rodriguez et al. 2009.6 Two reviewers (BD and AFP) worked independently at first to screen each record and each report retrieved, then reviewers discussed to achieve consensus. Any disagreement toward the included studies was resolved by consensus.
Two reviewers (BD and AFP) performed the literature selection and gathered the data from electronic databases that accumulated into a dataset qualified study that collected: the first author, the year of publication, the ethnicity, and patient-control proportions for CTLA-4 +49A/G genotyping into Microsoft Excel v2013. Using the distribution of the associated genotypes, the occurrence of the G-allele in CTLA-4 +49A/G were computed. The outcomes were compared, and differences were addressed via discussion.
Application of Review Manager version 5 (Cochrane Library, UK) was utilized to compute the available dataset from the included studies. The dataset included in the software was firstly collected in Microsoft Excel. Model of random effect was applied when p-value in heterogeneity test was <0.05. For dichotomous data, results were reported using OR, and 95% CI was also computed. Overall analysis needed p-value <0.05 to be interpreted as statistically significant. I2 was utilized for heterogeneity between the included studies as well.
This meta-analysis included 13 out of 15 papers7–21 investigating the relationship between CTLA-4 +49A/G SNPs and renal transplantation rejection risk. The data were extracted, and the occurrence of the G-allele of CTLA-4 +49A/G in the groups was determined. Table 1 presents the characteristics of the studies. These 13 studies included 959 patients with acute rejection and 2069 controls with non-acute rejection.
| Author, reference (year) | Case genotype | Control genotype | Country | Ethnicity | Genotyping | X2 HWE (control) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AA | AG | GG | N | AA | AG | GG | N | |||||
| Canossi et al.18 (2013) | 5 | 11 | 18 | 34 | 2 | 21 | 11 | 34 | Italy | Caucasians | Polymerase Chain Reaction | 3,6662 |
| Dmitrienko et al.7 (2005) | 3 | 29 | 18 | 50 | 3 | 24 | 23 | 50 | Canada | Caucasians | Polymerase Chain Reaction | 1,0204 |
| Domanski et al.16 (2012) | 13 | 35 | 22 | 70 | 32 | 96 | 51 | 179 | Poland | Caucasians | Polymerase Chain Reaction | 1,2887 |
| Gao et al.17 (2012) | 25 | 16 | 4 | 45 | 44 | 62 | 16 | 122 | China | Asians | Polymerase Chain Reaction | 0,6485 |
| Gendzekhadze et al.10 (2006) | 5 | 16 | 9 | 30 | 5 | 11 | 17 | 33 | Venezuela | Caucasians | Polymerase Chain Reaction | 1,7723 |
| Gorgi et al.9 (2006) | 14 | 10 | 7 | 31 | 22 | 15 | 2 | 39 | Tunisia | Africans | Polymerase Chain Reaction | 0,0745 |
| Kim et al.15 (2010) | 34 | 19 | 6 | 59 | 124 | 115 | 27 | 266 | Korea | Asians | Polymerase Chain Reaction | 0,0020 |
| Krichen et al.12 (2009) | 4 | 1 | 0 | 5 | 6 | 11 | 1 | 18 | Tunisia | Africans | Polymerase Chain Reaction | 1,8944 |
| Misra et al.19 (2014) | 10 | 12 | 14 | 36 | 20 | 56 | 78 | 154 | India | Asians | Polymerase Chain Reaction | 3,5822 |
| Ruhi et al.14 (2010) | 5 | 20 | 24 | 49 | 4 | 20 | 23 | 47 | Turkey | Caucasians | Polymerase Chain Reaction | 0,0141 |
| Ruhi et al.20 (2015) | 4 | 17 | 13 | 34 | 4 | 20 | 23 | 47 | Turkey | Caucasians | Polymerase Chain Reaction | 0,0141 |
| Slavcheva et al.21 2001 | 24 | 81 | 60 | 165 | 15 | 51 | 53 | 119 | USA | Mixed | Polymerase Chain Reaction | 0,2467 |
| Wisniewski et al.8 (2006) | 12 | 13 | 13 | 38 | 11 | 23 | 19 | 53 | Poland | Caucasians | Polymerase Chain Reaction | 0,6629 |
| Haimila et al.11 (2009) | 37 | 37 | 151 | 151 | Finland | Caucasians | Polymerase Chain Reaction | None | ||||
| Kusztal et al.13 (2010) | 22 | 22 | 48 | 48 | Poland | Caucasians | Polymerase Chain Reaction | None | ||||
As shown in Figure 2 and Table 2, in the overall analysis (OR=1.22, 95% CI:1.05-1.42, P=0.001), the CTLA-4 +49A/G G-allele was shown to be related to an increased risk of acute rejection after renal transplantation in this meta-analysis. In contrast, the A-allele seemed to be related with protective effect toward acute rejection risk. For renal transplantation, the presence of the GG-genotype (OR=1.47, 95% CI:1.14-1.89, P=0.003) increased the risk of acute rejection, as shown in Figure 3 and Table 2. However, the AA-genotype did not seem to protect against the acute rejection associated with transplantations (OR=0.87, 95% CI:0.69-1.09, P=0.21). This meta-analysis found that the CTLA-4 +49A/G SNPs were not linked to an increased risk of acute rejection in African or Caucasian ethnicities, according to multilevel modelling of analyses (Table 2). However, the CTLA-4 +49A/G G-allele was related with renal transplantation rejection risk among Asian populations, as shown in Figure 4 (OR=1.55, 95% CI:1.16-2.06, P=0.003; Table 2), but not in the general population. The GG-genotype was also linked to an increased risk of acute rejection after the analysis, as shown in Figure 5 (OR=1.91, 95% CI:1.29-2.84, P=0.001) and Table 2.
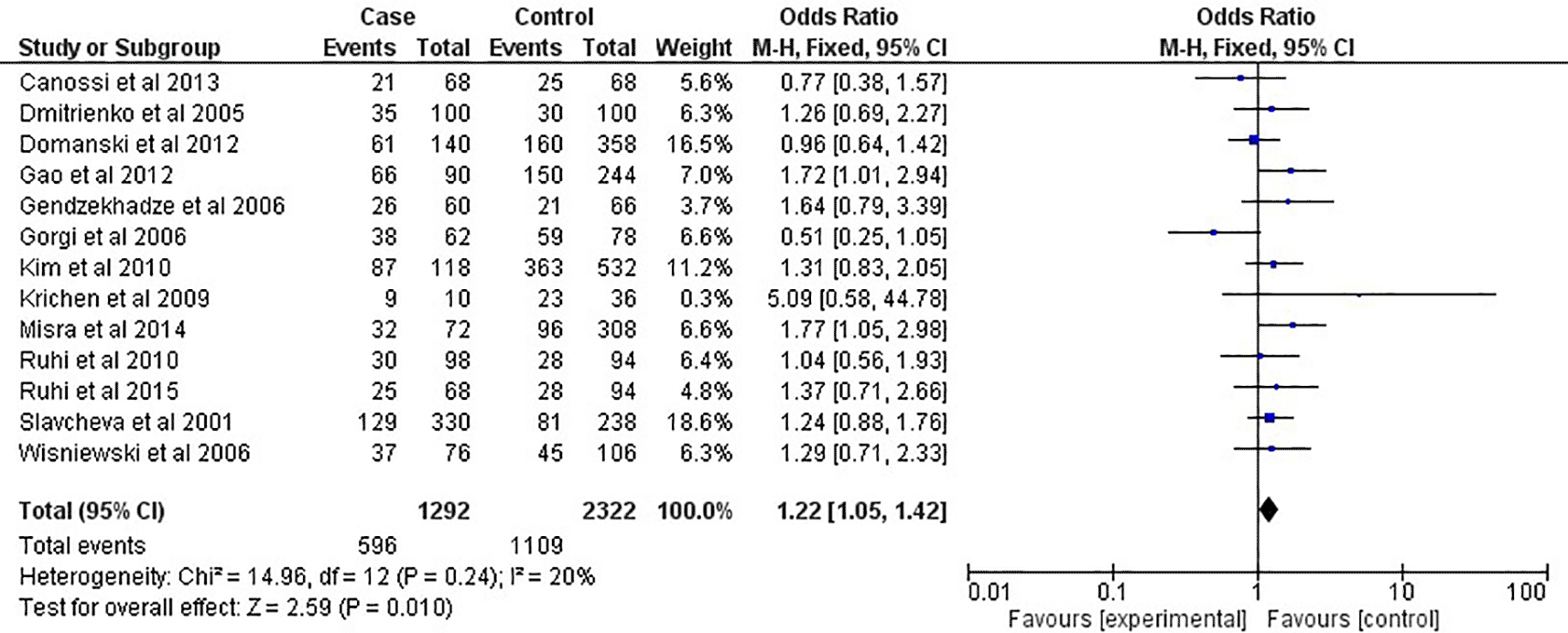
CI: confidence interval; P: P-value; I2: heterogeneity.
| Allele & genotype | NS | Model | Sensitivity, % | Specificity, % | OR | 95%CI | pH (Tau2 if random) | I2 | P |
|---|---|---|---|---|---|---|---|---|---|
| Overall analysis | |||||||||
| G vs. A | 13 | Fixed | 50.64 | 44.56 | 1.22 | 1.05 – 1.42 | 0.24 | 20% | 0.010* |
| A vs. G | 13 | Fixed | 49.36 | 55.40 | 0.82 | 0.71 – 0.95 | 0.24 | 20% | 0.010* |
| GG vs. AG+AA | 13 | Fixed | 30.78 | 63.90 | 1.47 | 1.14 – 1.89 | 0.60 | 0% | 0.003* |
| AG vs. AA+GG | 13 | Fixed | 39.72 | 61.40 | 0.88 | 0.29 – 1.61 | 0.10 | 36% | 0.22 |
| AA vs. AG+GG | 13 | Fixed | 29.50 | 74.71 | 0.87 | 0.69 – 1.09 | 0.28 | 17% | 0.21 |
| Asian sub-group | |||||||||
| G vs. A | 3 | Fixed | 66.07 | 43.82 | 1.55 | 1.16 – 2.06 | 0.62 | 0% | 0.003* |
| A vs. G | 3 | Fixed | 33.93 | 56.18 | 0.65 | 0.48 – 0.86 | 0.62 | 0% | 0.003* |
| GG vs. AG+AA | 3 | Fixed | 49.29 | 65.31 | 1.91 | 1.29 – 2.84 | 0.57 | 0% | 0.001* |
| AG vs. AA+GG | 3 | Fixed | 33.57 | 57.01 | 0.65 | 0.44 – 0.96 | 0.64 | 0% | 0.03 |
| AA vs. AG+GG | 3 | Fixed | 17.14 | 77.68 | 0.72 | 0.43 – 1.22 | 0.71 | 0% | 0.22 |
| Caucasian sub-group | |||||||||
| G vs. A | 7 | Fixed | 48.49 | 42.76 | 1.12 | 0.90 – 1.39 | 0.74 | 0% | 0.32 |
| A vs. G | 7 | Fixed | 51.51 | 57.24 | 0.90 | 0.72 – 1.11 | 0.74 | 0% | 0.32 |
| GG vs. AG+AA | 7 | Fixed | 29.12 | 59.50 | 1.31 | 0.85 – 2.01 | 0.95 | 0% | 0.22 |
| AG vs. AA+GG | 7 | Fixed | 38.74 | 66.51 | 0.95 | 0.71 – 1.28 | 0.10 | 44% | 0.74 |
| AA vs. AG+GG | 7 | Fixed | 32.14 | 73.99 | 0.92 | 0.68 – 1.25 | 0.26 | 23% | 0.60 |
| African sub-group | |||||||||
| G vs. A | 2 | Random | 65.28 | 28.07 | 1.28 | 0.14 – 12.04 | 0.05 (2.04) | 75% | 0.83 |
| A vs. G | 2 | Random | 34.72 | 71.93 | 0.78 | 0.08 – 7.36 | 0.05 (2.04) | 75% | 0.83 |
| GG vs. AG+AA | 2 | Random | 50 | 50.88 | 1.76 | 0.15 – 20.33 | 0.05 (2.37) | 73% | 0.65 |
| AG vs. AA+GG | 2 | Fixed | 30.56 | 54.39 | 0.58 | 0.24 – 1.42 | 0.23 | 29% | 0.23 |
| AA vs. AG+GG | 2 | Fixed | 19.44 | 94.74 | 3.99 | 0.99 – 16.10 | 0.39 | 0% | 0.05 |
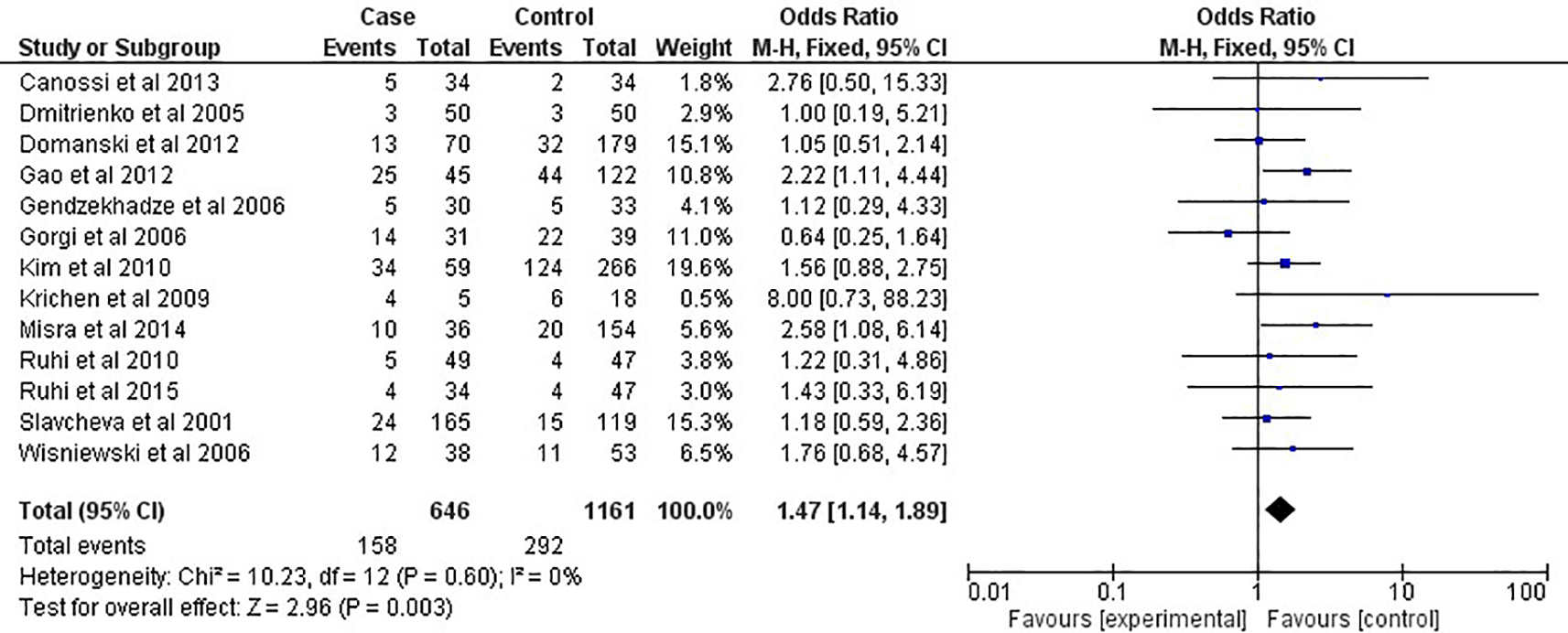
CI: confidence interval; P: P-value; I2: heterogeneity.
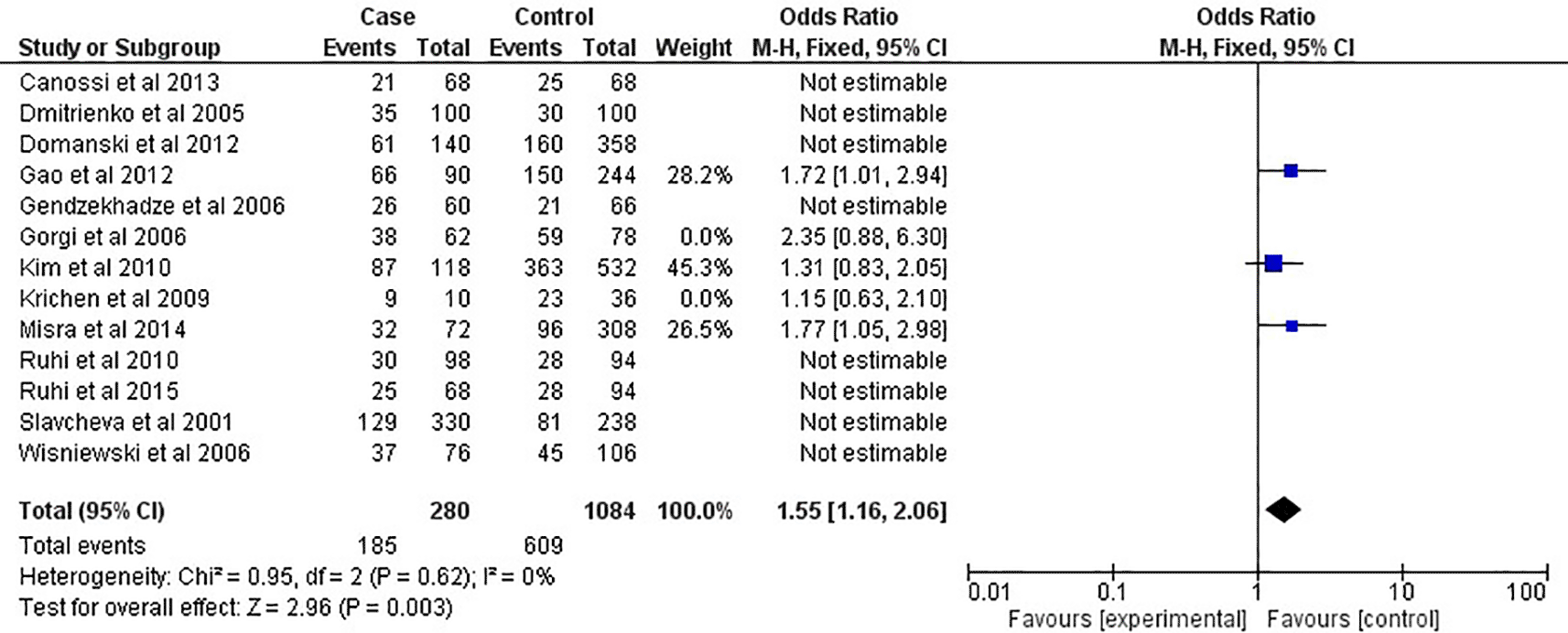
CI: confidence interval; P: P-value; I2: heterogeneity.
This meta-analysis found a connection between the CTLA-4 +49A/G G-allele/GG-genotype and the increased prevalence of acute renal transplantation rejection, with more prominent emphasis of evolution influence exclusion in the genetic polymorphism study. Additionally, we tested for publication bias and found that the CTLA-4 +49A/G SNPs had no influence on an increased incidence of acute renal rejection in general populations. The association of CTLA-4 +49A/G SNPs and acute renal rejection risk was shown to be strong. After further investigation, we discovered no association between the CTLA-4 +49A/G SNPs and the incidence of the rejection in the African and Caucasian population. Caucasians and Africans were underrepresented in the included studies. Therefore, further research about the role of the polymorphism among different ethnicities is necessary to be done.
Intriguingly, we discovered that the G-allele and GG-genotype were both linked to risk of renal transplantation rejection in Asians. It means that the G-allele and GG-genotype seemed to be a risk factor for acute rejection in Asian populations, according to this study. However, further research is needed to examine the link between the two variables. Our findings seemed to be quite stable in certain respects. Meta-analysis studied by Duan et al.22 found that the G-allele was related with a higher risk of acute rejection after renal transplantation, but not with the GG-genotype and the AA-genotype in the general population. However, a limitation of the previous paper was not classifying cases into ethnic subgroups, also the previous publication was not validated by the Hardy-Weinberg equilibrium that explains evolution theory influences that need to be excluded in order to make sure the SNPs occurred actually came from the patient’s case itself.
According to Zhu et al.,23 the CTLA-4 +49A/G SNPs were not related with the incidence of renal transplant rejection in general populations. In a meta-analysis of nine studies, Gao et al.24 found that the G-allele and GG-genotype were related with acute renal rejection risks in Asian populations and in general populations. As a consequence of the increased number of studies included in our meta-analysis (13 included studies), our findings were more vigorously updated than those from the prior meta-analyses, also the use of HWE analysis provides more valid results in terms of polymorphism study. CTLA-4 +49A/G SNPs, particularly the G-allele/GG-genotype, were associated with acute renal rejection risk in Asian ethnicity and overall populations, according to our findings. Because of a wide range of issues including linguistic bias, a small sample size, poor statistical power and heterogeneity of recruited patients, as well as a variety of research designs and varied therapies, the findings should be taken with a grain of salt.
To summarize, the findings of the conducted meta-analysis indicate that the CTLA-4 +49A/G SNPs, particularly the G-allele/GG-genotype, were related with the acute rejection, increased risk in renal transplantation cases of Asian ethnicity and global populations. But in spite of that, further association research is necessary to better elucidate this.
Figshare: PRISMA checklist and flow diagram for ‘Cytotoxic T-lymphocyte associated-protein-4 +49A/G-allele (rs231775) Single Nucleotide Polymorphisms was corresponded with acute allograft renal transplantation rejection: a multilevel modelling of meta-analysis’. https://doi.org/10.6084/m9.figshare.20124215.25
Data are available under the terms of the Creative Commons Attribution 4.0 International license (CC-BY 4.0).
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Are the rationale for, and objectives of, the Systematic Review clearly stated?
Yes
Are sufficient details of the methods and analysis provided to allow replication by others?
Yes
Is the statistical analysis and its interpretation appropriate?
Yes
Are the conclusions drawn adequately supported by the results presented in the review?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Medicine
Are the rationale for, and objectives of, the Systematic Review clearly stated?
Yes
Are sufficient details of the methods and analysis provided to allow replication by others?
Yes
Is the statistical analysis and its interpretation appropriate?
I cannot comment. A qualified statistician is required.
Are the conclusions drawn adequately supported by the results presented in the review?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: kidney transplant sorgen , ABO incompatible kidney transplantation
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 05 Aug 22 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)