Keywords
mass spectrometric imaging, interactive analysis, clustering, labeling
This article is included in the Bioinformatics gateway.
mass spectrometric imaging, interactive analysis, clustering, labeling
Mass spectrometry imaging (MSI) is a technique of building a map of the spatial distribution of molecular features across the tissue of interest without pretreatment.1–4 This technique is a promising complement to the gold standard of tissue analysis – histology study that is a time-consuming, labor-intensive, and sometimes subjective method.
Each pixel of raw MSI-map is a mass spectrum of the corresponding location in the sample. This multidimensional map is almost impossible to interpret by naked eyes and has to be converted to a simpler representation for better visualization and usability.5–12
In our previous work, we introduced13 a fast, precise, and accurate imaging tool based on the interactive building of the cosine similarity measure maps (CSMM) between the reference pixel and the rest of the image. Introduced technique well suited for visual estimation of presence, location, and level of heterogeneity of homogeneous regions in the image. It also allows extraction of the region reference mass spectra, and evaluation of the influence of the reference pixels on the distribution of similarity characteristics on the map.
Here we present a user-friendly interface for building and analysis of CSMMs of MSI data.
Here we introduce Interactive CSMM, a MATLAB app, which provides an intuitive graphical interface for interactive evaluation of mass spectrometric imaging heterogeneity. An example of the interface is shown in Figure 1. Interactive CSMM was created in MATLAB R2019b. We also introduce the Python script for converting standard raw imzML file format to mat-file, which is required by the Interactive CSMM. The script currently depends on the following libraries: numpy, psutil, pyimzML and scipy.
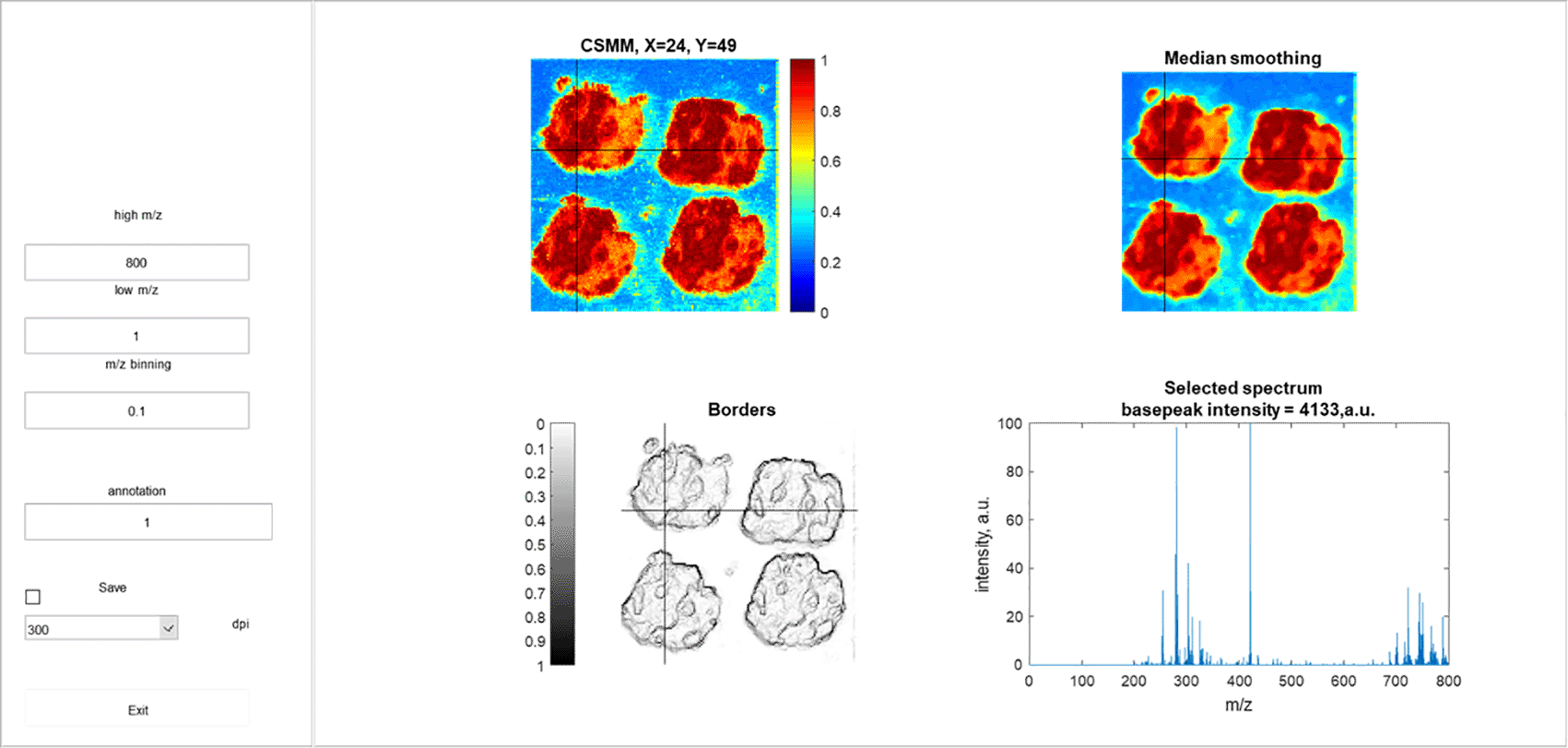
On the right panel on the top left CSMM is located, on the top right the result of CSMM median smoothing is displayed, on the bottom left is a map of the boundaries of homogeneous regions, and the mass spectrum of the reference pixel is shown on the bottom right.
The Interactive CSMM can be launched locally from any computer with MATLAB (R2019b or higher; lower versions also might work properly) installed. Installation and launching instructions are also available. All interfaces and plots of Interactive CSMM are highly interactive, allowing users to visualize data in real-time with interactive selection reference mass spectrum, as well as store the results of the analysis.
The program 1) allows the user to interactively select the reference pixel, specify the mass range and other parameters for data binning 2) calculates the CSMM of all imaging data with respect to the selected pixel 3) label homogeneous areas and save the assigned area number, coordinates of the reference pixel and the name of the CSMM image file built on this pixel to a text file 4) save publication-ready images of CSMM with specified resolution.
To improve the interpretability of the image and clean up the pixelation, outliers, and measurement artifacts the program provides the smoothed version of CSMM, the visual map of the boundaries of homogeneous zones,13 and the mass spectrum of the reference pixel.
The presence in the spectra ions distributed over the tissue equally could cause blurring of the picture. So, we provide users with options to specify ranges is m/z which are reflecting the tissue heterogeneity, and compare positions and shape of homogeneity regions obtained in different m/z ranges.
Another effect that complicates the interpretation of MS images is the presence of transition zones due to gradual changes in the ion’s intensity within such zones. In our method, due to the building of the CSMM over the mass range, the transition zones become clearly visible because they include the peaks of both boundary zones. That effect is more difficult to achieve with standard imaging approaches (Supplementary Materials of the method describing article13), which consider the distribution of individual ions.
The additional benefit of our method is that there is no data preprocessing is required other than binning. The parameters of binding can be changed online. No alignment is required as well, it can be replaced with a larger binning. It could be also shown that the method works well with non-normalized data and there is no need for baseline correction.
CSMM allows you to define zones of least and greatest similarity. It does not automatically divide the measured map into zones. But you can find areas with similar spectra by varying the reference pixel. By changing the other parameters (to a greater extent, the mass range), it is possible to optimize the CSMM color map and improve the visualization of heterogeneity of the measured sample. The smooth changes in heterogeneity can be observed as smooth color changes on the CSMM.
We tested this method on different data sources14 (measured with different ion sources: MALDI-imaging, DESI-imaging; and different mass analyzers TOF MS, Orbitrap, ICR MS) (Supplementary Materials of the method describing article13). Our article presents CSMMs for colorectal adenocarcinoma data.
Preprocessing steps and descriptions of operations are presented in the manual.
We presented software that allows users to quickly evaluate the presence and structure of heterogeneous areas in the sample, and manually make a dataset of feature spectrum for the homogeneous zones. By varying the reference spectrum and the mass range used in the construction of the CSMM, it is possible to understand which mass ranges most reflect the heterogeneity and make the boundaries between the zones more contrasting. A more detailed discussion is presented in the method describing article.13
GigaDB: Supporting materials for “Benchmark datasets for 3D MALDI- and DESI-Imaging Mass Spectrometry”, http://dx.doi.org/10.5524/100131.14
This project contains the following underlying data:
- 3DMouseKidney.ibd
- 3DMouseKidney.imzML
- 3D_Mouse_Pancreas.ibd
- 3D_Mouse_Pancreas.imzML
- 3D_OSCC.ibd
- 3D_OSCC.imzML
- ColAd_Individual.zip
- Colorectal_Adenocarcinoma.h5
- Microbe_Interaction_3D_Timecourse_LP.ibd
- Microbe_Interaction_3D_Timecourse_LP.imzML
Data are available under the terms of the Creative Commons Zero “No rights reserved” data waiver (CC0 1.0 Public domain dedication).
Data are also available at Metabolights: MTBLS176: Benchmark datasets for 3D MALDI- and DESI-Imaging Mass Spectrometry.
Source code available from: https://github.com/EvgenyZhvansky/Interactive_CSMM/.
Archived source code at time of publication: http://doi.org/10.5281/zenodo.5776541.
License: Creative Commons Attribution 4.0 International license (CC-BY 4.0).
Conceptualization A.S., E.S.Z.; methodology A.S., E.S.Z.; software E.S.Z.; investigation M.B., E.V.Z; writing — original draft preparation E.S.Z., E.V.Z., M.S.; writing — review and editing E.S.Z., E.V.Z., A.S., K.B.; visualization E.S.Z.; supervision A.S.; project administration A.S.; funding acquisition A.S.; formal Analysis E.S.Z.; data curation M.B., M.S., S.S., K.B.; validation E.V.Z., M.B.; resources M.B., E.V.Z. All authors have read and agreed to the current version of the manuscript.
The research used the equipment of Shared Research Facilities of N.N. Semenov Federal Research Center for Chemical Physics of the Russian Academy of Sciences.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the rationale for developing the new software tool clearly explained?
Partly
Is the description of the software tool technically sound?
Yes
Are sufficient details of the code, methods and analysis (if applicable) provided to allow replication of the software development and its use by others?
Yes
Is sufficient information provided to allow interpretation of the expected output datasets and any results generated using the tool?
Partly
Are the conclusions about the tool and its performance adequately supported by the findings presented in the article?
Yes
References
1. Zhvansky ES, Ivanov DG, Sorokin AA, Bugrova AE, et al.: Interactive Estimation of Heterogeneity from Mass Spectrometry Imaging.Anal Chem. 2021; 93 (8): 3706-3709 PubMed Abstract | Publisher Full TextCompeting Interests: No competing interests were disclosed.
Reviewer Expertise: Mass Spectrometry Imaging
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |
|---|---|
| 1 | |
|
Version 1 25 Jan 22 |
read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)