Keywords
Annelida, Syllidae, Syllis, Mitochondrial genome
This article is included in the Genomics and Genetics gateway.
Annelida, Syllidae, Syllis, Mitochondrial genome
Mitochondria are cellular organelles that function as an energy factory in animals. They are inherited from the maternal lineage and can be used to trace phylogenetical relationships. The mitochondrial genome sequence thus provides very valuable data for genetic or taxological research. In addition, the sequence and order of genes in the mitochondrial DNA (mtDNA) can be used to uncover phylogenetic relationships between evolutionarily close or distant species (Vallès and Boore 2006). Owing to the startling progress of sequencing technology leading to next-generation sequencing (NGS), many mitogenome sequences from diverse species have accumulated in public databases in recent years. However, obtaining complete mitogenome data remains difficult because of the diversity of mitogenome structure, gene arrangement, and transfer RNA (tRNA) structure, according to species (Bernt et al. 2013).
More than 700 species of the family Syllidae have been classified into 74 genera (Aguado and Martín 2009). Despite the number of species, the taxonomy of the family Syllidae remains incomplete with many unclarified correlations. Few studies describing the mtDNA sequence of members of the phylum Annelida are available from the first decade of the 2000s (Boore 2004; Jennings and Halanych 2005; Bleidorn et al. 2006; Zhong et al. 2008; Mwinyi et al. 2009; Shen et al. 2009). However, some mtDNA sequences from the phylum Annelida were reported using NGS (Aguado et al. 2016), which was revealed that the gene order in the mtDNA sequence in this phylum is well conserved, although Weigert et al. (2016) reported that differences in gene order occur more frequently than expected. Despite this controversy, information on the mtDNA sequences of the phylum Annelida remains insufficient. In this study, we obtained the complete mitochondrial genome sequence of a novel species belonging to the genus Syllis. This mitogenome contributes to our understanding of the conservation of mitochondrial gene order in the phylum Annelida.
The molecular biology experiments were conducted at Pukyong National University. The specimen was obtained (36°12′N, 129°25′E) from a small worm attached to Farrer’s scallop (Chlamys farreri K. H. Jones & Preston, 1904) and this small worm had a lots of legs on the body. Genomic DNA (gDNA) was extracted from the whole body of the worm using the Bead™ Genomic DNA Prep Kit For Animal Tissue (Biofact, Republic of Korea). We deposited the gDNA at Pukyong National University (Voucher no. PKNU_2021_002: Jun Young Chae, jychae@pukyong.ac.kr) because the whole worm was used for extraction gDNA owing to its very small size. The cox1 gene was amplified using an invertebrate universal primer set (LCO1490, 5′-GGTCAACAAATCATAAAGATATTGG-3′; HCO2198, 5′-TAAACTTCA-GGGTGACCAAAAAATCA-3′; Folmer et al. 1994) by PCR. PCR was conducted using HelixAmp™ Taq-Plus Polymerase (NANOHELIX, Republic of Korea) and SimpliAmp™ Thermal Cycler (RRID:SCR_023004). The PCR condition was pre-denaturation at 95°C for 2 minutes; 30 cycles at 95°C for 20 seconds, 40°C for 40 seconds, and 72°C for 45 seconds; final extension at 72°C for 5 minutes. The amplicon was sequenced, and the cox1 sequence was compared to identify this species using the NCBI Basic Local Alignment Search Tool (BLAST) (RRID:SCR_004870) (Johnson et al. 2008).
The bioinformatics experiments were conducted at theMOAGEN. gDNA (1 μg) was sheared using the S220 Ultra sonicator (Covaris, USA). MGIEasy DNA library prep kit (MGI, China) was used for library preparation according to the manufacturer’s instructions. Briefly, fragmented gDNA was selected based on its size using AMPure XP magnetic beads and the fragmented gDNA was end-repaired and a-tailed at 37°C for 30 minutes, and 65°C for 15 minutes. Indexing adapter was ligated to the ends of the DNA fragments at 23°C for 60 minutes. PCR was performed to enrich those DNA fragments that have adapter molecules after purifying the adapter-ligated DNA. Thermocycler conditions were as follows: 95°C for 3 minutes, 7 cycles of 98°C for 20 seconds, 60°C for 15 seconds, and 72°C for 30 seconds, with a final extension at 72°C for 10 minutes. The double stranded library is quantified using QauntiFluor ONE dsDNA System (Promega, USA). The library is circularized at 37 °C for 30 minutes, and then digested at 37°C for 30 minutes, followed by cleanup of circularization product. The library is incubated at 30°C for 25 minutes using DNB enzyme for making DNA nanoball. Finally, the library was quantified by QauntiFluor ssDNA System (Promega, USA). NGS was conducted on the MGISEQ-2000 (MGI, China) platform with 150 bp paired-end reads. The raw reads were screened using the cutadapt tool (RRID:SCR_011841) (Martin 2011), and all clean sequences were used for de novo assembly using the assembler of the CLC Genomics Workbench (RRID:SCR_011853) ver. 20.04 (Qiagen). The circular form of the mitogenome was confirmed using the “Map to Reference” tool of Geneious (RRID:SCR_010519) software ver. 2021.2.2 by remapping the filtered data into the contig sequence from the de novo assembly. Annotation of the complete mtDNA sequence was performed using the MITOS WebServer and manually corrected using SnapGene (RRID:SCR_015052) software ver. 5.3.2 based on previously released mtDNA annotation information (Aguado et al. 2016). The mitogenome map was prepared using ORDRWA (Greiner et al. 2019). We also obtained a contig that included the 18S rRNA sequence from the de novo assembly of CLC Genomics Workbench, registered at GenBank (accession no. OP341337) (Chae and Kim 2022b), and used this sequence to construct a phylogenetic tree for a more detailed taxonomic classification of this worm.
Bayesian inference (BI) with MrBayes (RRID:SCR_012067) 3.2.6 (Huelsenbeck and Ronquist 2001), was used to perform phylogenetic analysis based on nucleotide sequences of 13 protein-coding genes (PCGs) of 16 available mitochondrial genomes in the class Polychaeta, and Orbinia latreilliid (accession no. NC_007933) (Bleidorn et al. 2006) was set as an outgroup. Additional phylogenetic analysis was conducted based on the nucleotide sequences of 18S rRNA of 41 species belonging to the family Syllidae.
BLASTN analysis showed that the partial sequence of cox1 from the worm had the highest similarity to that of Syllis pigmentata (accession no. EF123774.1), at 87.76%, which is a relatively low identity score. The sequence was also similar to that of S. ehlersioides (accession no. EF123773.1), S. alternat (accession no. HQ932467.1), S. albae (accession no. KX792209.1), Typosyllis antoni (accession no. KX752426.1), and T. augeneri (accession no. JF903788.1) in decreasing order at 87.62%, 83.07%, 82.76%, 81.90%, and 77.70%, respectively. All these species belong to the genus Syllis, suggesting that our worm may be a new species belonging to this genus.
The raw data output of NGS was deposited in the Sequence Read Archive (SRA) database (accession no. SRR18465399) (theMOAGEN 2022b). The length of the complete mtDNA sequence was 17,092 bp, and the sequence was registered in GenBank (accession no. ON312495) (Chae and Kim 2022a). A total of 38 genes were predicted in this mitochondrial genome, including 13 PCGs, 23 tRNA genes, and two rRNA genes, and all genes are encoded on the positive strand. The gene composition of this mtDNA is similar to that of T. antoni (Aguado et al. 2016), with two tRNA-M being present and consistent with previously reported data (Kurabayashi et al. 2006; Zhang et al. 2009).
Almost all PCGs start with an ATG codon (atp6, cox3, nad2, cytb, atp8, cob, nad3, nad1, cox2, nad4, and nad4l), whereas nad6 and nad5 have ATA as a start codon and cox1 uniquely starts with an AAC codon found in insects (Kim et al. 2016). Nad2 and nad1 terminate with truncated T- codons. The remaining 11 PCGs have a TAA stop codon, except for nad4 and nad4l, which have TAG stop codons. The mtDNA of T. antoni contains cox3 as the first gene and tRNA-P as the last gene. Although the worm mtDNA examined in this study begins with tRNA-G and ends with tRNA-W, the gene order is the same from nad6 to cox2 (nad6, tRNA-F, tRNA-D, tRNA-T, tRNA-S2, tRNA-K, tRNA-Y, Large rRNA, tRNA-R, tRNA-S1, tRNA-E, tRNA-V, tRNA-I, atp8, cob, nad3, tRNA-N, tRNA-M, tRNA-M, nad5, nad1, and cox2) in the two species (Figure 1). By contrast, the mitogenome sequences of Ramisyllis multicaudata and R. kingghidorahi are entirely dissimilar from that of the worm described in this study. Their gene order is the same because these two species belong to the same genus. Similarly, the differences in gene order between the mitogenomes of Syllis sp. and T. antoni result from these species being from different genera. These results are consistent with gene order in mitogenomes being more diverse than expected and accord with previous reports of different gene orders in the Phylum Annelida (Aguado et al. 2016; Weigert et al. 2016). We identified 23 tRNA genes, including two tRNA-L, two tRNA-S, and two tRNA-M. The standard cloverleaf structure was observed in the predicted secondary structure of 15 tRNAs, except for tRNA-R and tRNA-S2, which lack a D-arm, and five tRNAs (tRNA-D, tRNA-F, tRNA-G, tRNA-M, and tRNA-Y), which lack a loop structure in the T-arm. Small rRNA with 880 bp was located between tRNA-L1 and tRNA-A. Large rRNA was observed between tRNA-Y and nad2, and its length was 1016 bp. The length of the putative control region was 1,291 bp and was located between tRNA-W and tRNA-G. The gene order is different from that of mitogenomes of other species in the family Syllidae (Figure 1).
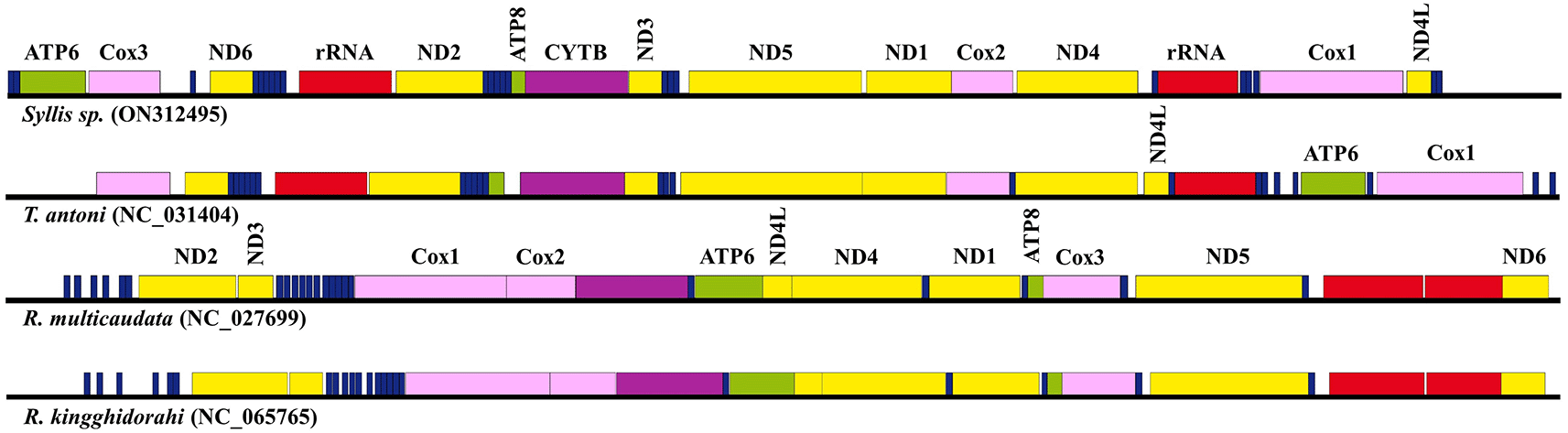
The mitogenome gene order of the Syllis sp. described in this study was compared with that of other species belonging to the family Syllidae. The circular mitogenomes in these mtDNA maps are shown linearly to more easily compare the gene orders. The tRNAs, ATP synthase F0 subunits, cytochrome c oxidase subunits, NADH dehydrogenase subunits, ribosomal RNAs, and cytochrome b are marked blue, green, pink, yellow, red, and purple, respectively. The gene name is indicated at the top of each box. mtDNA, mitochondrial DNA; tRNA, transfer RNA.
Phylogenetic analysis showed that the Syllis sp. was clustered with T. Antoni, and the clade had a close relationship with the family Syllidae members belonging to the order Phyllodocida (Figure 2), suggesting that this worm is a previously unclassified species. Additional phylogenetic analysis was performed to confirm the genus of the worm based on the 18S rRNA sequence. The results show that it was grouped with S. busseltonensis, and this node was assembled with various Syllis spp. rather than with the genus Haplosyllis or Branchiosyllis (Figure 3). These results are consistent with the examined worm being a novel Syllis sp. that has not yet been described.

A phylogenetic tree was constructed using the nucleotide sequence of PCGs of the Syllis sp. described in this study and 16 species obtained from GenBank with Orbinia latreilliid (NC_007933) as an outgroup. GenBank accession numbers are given with species names. Posterior probabilities of the Bayesian inference are indicated as node numbers. Class, order, and family taxonomic ranks are shown adjacent to vertical black bars. The Syllis sp. analyzed in this study is indicated by an arrowhead. PCG, protein-coding genes.
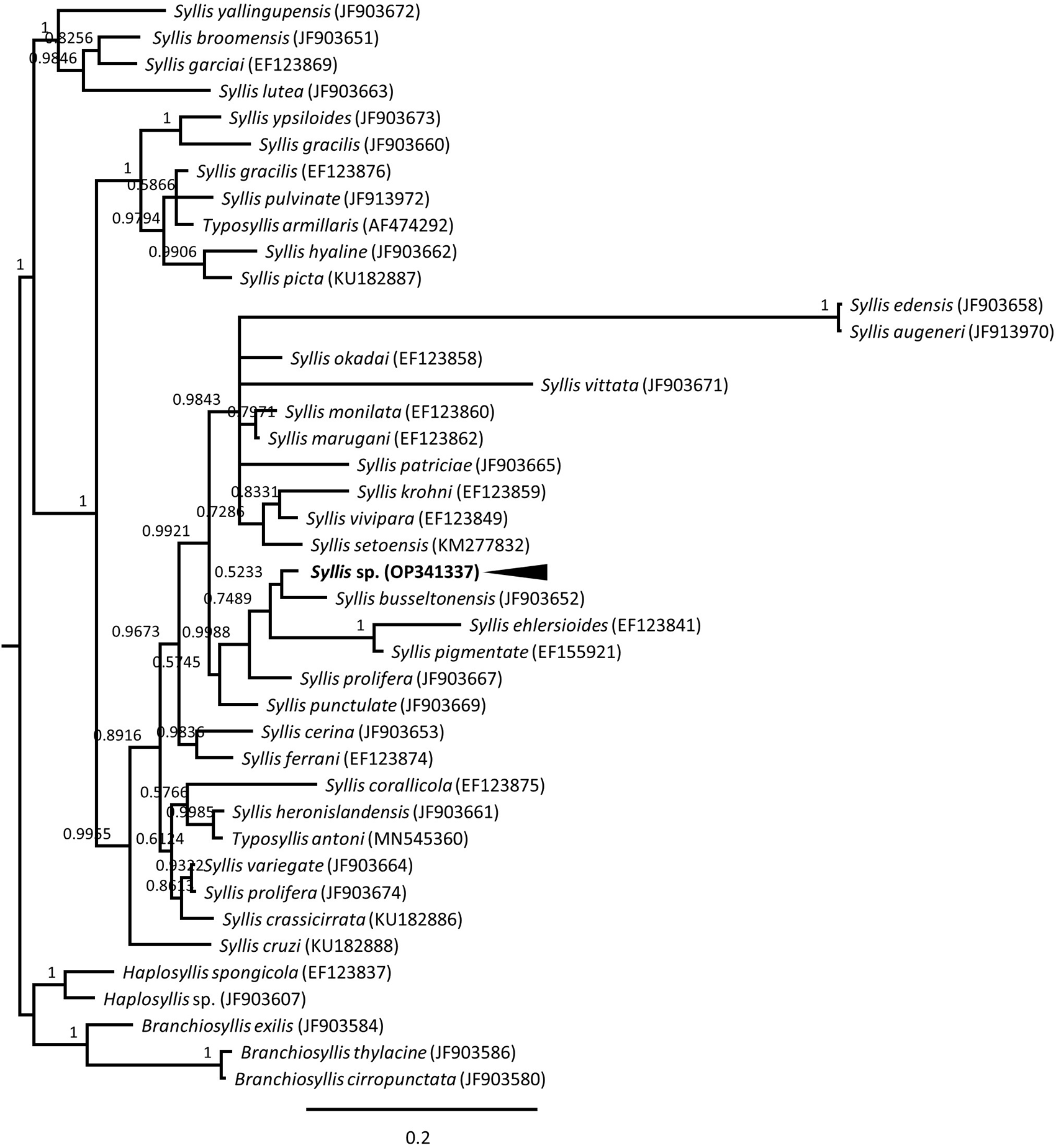
A phylogenetic tree was constructed based on the nucleotide sequence of the 18S rRNA gene from the Syllis sp. described in this study and the nucleotide sequences of 40 species from the family Syllidae in GenBank. GenBank accession numbers accompany species names. Node numbers indicate the posterior probabilities of the Bayesian inference. The Syllis sp. described in this study is indicated by the arrowhead. rDNA, ribosomal DNA.
There is no human or animal involvement in the study. Since the sample is an insect, there is no need for ethical approval or permission to collect the sample.
GenBank: Syllis sp. JYC-2022 mitochondrion, complete genome. Accession number ON312495; https://identifiers.org/ncbi/insdc:ON312495 (Chae and Kim 2022a).
GenBank: Syllis sp. JYC-2022 small subunit ribosomal RNA gene, partial sequence. Accession number OP341337; https://identifiers.org/ncbi/insdc:OP341337 (Chae and Kim 2022b).
BioProject: Syllis sp. JYC-2022. Accession number PRJNA818342; https://identifiers.org/NCBI/bioproject:PRJNA818342 (theMOAGEN 2022a).
SRA: Syllis sp. JYC-2022. Accession number SRR18465399; https://identifiers.org/insdc.sra:SRR18465399 (theMOAGEN 2022b).
BioSample: Syllis sp. JYC-2022. Accession number SAMN26856052; https://identifiers.org/biosample:SAMN26856052 (theMOAGEN 2022c).
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Are the rationale for sequencing the genome and the species significance clearly described?
Yes
Are the protocols appropriate and is the work technically sound?
Yes
Are sufficient details of the sequencing and extraction, software used, and materials provided to allow replication by others?
Yes
Are the datasets clearly presented in a usable and accessible format, and the assembly and annotation available in an appropriate subject-specific repository?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Taxonomy and phylogeny of Annelida, Biodiversity and Ecology of marine benthos
Are the rationale for sequencing the genome and the species significance clearly described?
Partly
Are the protocols appropriate and is the work technically sound?
No
Are sufficient details of the sequencing and extraction, software used, and materials provided to allow replication by others?
Partly
Are the datasets clearly presented in a usable and accessible format, and the assembly and annotation available in an appropriate subject-specific repository?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Hydrobiology
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 31 Aug 23 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)