Keywords
Glaucoma, Classification, AlexNet, Convolutional Neural Network (CNN), Diabetic Retinopathy
This article is included in the Artificial Intelligence and Machine Learning gateway.
This article is included in the AI in Medicine and Healthcare collection.
Glaucoma, Classification, AlexNet, Convolutional Neural Network (CNN), Diabetic Retinopathy
The leading causes of blindness and poor vision around the globe are primarily age-related eye diseases such as glaucoma and diabetic retinopathy (DR).1–4 Glaucoma is a condition caused by elevated intraocular pressure.1 The most common are open-angle glaucoma, angle-closure glaucoma, normal-tension glaucoma, and congenital glaucoma.2 On the other hand, DR is the most frequent complication of diabetes mellitus.3 It occurs because the small blood vessels in the retina swell and bleed or leak fluid, causing retinal damage and vision problems.3,4 DR has five stages or classes: normal, mild, moderate, severe and proliferative DR.4
Ophthalmic examination is essential for the diagnosis of glaucoma and DR. The following tests are carried out by physicians in order to perform a diagnosis for glaucoma: measuring intraocular pressure (tonometry),5 analyzing optic nerve damage with a dilated eye exam, checking areas of vision loss (visual field test),6 measuring corneal thickness (pachymetry)7 and inspecting the angle of drainage (gonioscopy).8 As most of these are imaging tests of the eye, it is essential to have accurate high quality images in order to perform a correct diagnosis of the disease. Similarly, DR is usually detected by physicians through comprehensive ophthalmologic examinations with dilation: taking cross-sectional images that show the thickness of the retina where fluid may be leaking from damaged blood vessels (optical coherence tomography)9 and injecting a special dye that place blood vessels with blockages and blood vessels leaking blood (fluorescein angiography).10 The diagnoses of these diseases require highly qualified medical personnel, therefore, expensive in terms of time and costs.
The artificial intelligence (AI) through deep learning methods provides computers with the ability to identify patterns in large image datasets and make predictions (predictive analysis).11–14 As a consequence, it is important to use AI as a tool to automatically analyze the fundus images and assist the physicians. This results in accessible, reliable, and affordable detection of glaucoma and other pathologies in general that affect vision (Table 1).
| Purpose | Method | Database (size) | Number of classes | Performance measure | Ref. | |||
|---|---|---|---|---|---|---|---|---|
| Accuracy (%) | Sensitivity (%) | Specificity (%) | AUC | |||||
| Diabetic retinopathy detection and classification | Fused CNN512, CNN299, and CNN (YOLOv3, EfficientNetB0) | DDR (13673), and Kaggle (3662) | 5 | 89.00 | 89.00 | 97.30 | 0.97 | 15 |
| Diabetic retinopathy detection and classification | GoogleNet | Kaggle (200) | 5 | 88.00 | 75.00 | 52.00 | - | 16 |
| Automated Identification of Diabetic Retinopathy | Customized deep CNN | EyePacs, MESSIDOR 2 and E-Ophtha (75137) | 2 | - | 74.00-94.00 | 80.00-98.00 | 0.94-0.97 | 17 |
| Diabetic retinopathy detection and classification | CNN (modified AlexNet) | Messidor (1190) | 4 | 95.60-96.60 | 88.00-96.00 | 97.30-96.60 | - | 18 |
| Classification of cataract fundus image | CNN (five layers) | (7851) | 2 to 4 | 90.82-94.07 | - | - | - | 19 |
| Cataract diagnosis and grade | CNN (ResNet-18) | (1352) | 6 | 92.66 | - | - | - | 20 |
| Glaucoma detection | CNN (LeNet & U-net) | RIM-ONE, DRISHTI-GS, DRIONS-DB, JSIEC, NIO and DRIVE | 2 | 98.8 | - | 99 | - | 21 |
| Glaucoma detection | CNN | CGSA (269601) | 3 | - | 82.2 | 70.4 | 0.82 | 22 |
Related to AI, convolutional neuronal networks (CNNs) are a class of deep learning method, most commonly applied to analyze visual imagery. Generally, they are made of a key set of basic layers, including the convolution layer, the sub-sampling layer, dense layers, and the soft-max layer.15–22
Among the different CNNs, AlexNet by Krizhevsky et al. achieved a new state-of-the-art recognition accuracy against all the traditional machine learning and computer vision approaches that offer the opportunity to be retrained.23 AlexNet is structured with eight main layers, which consists of five convolutional layers (with max pooling after the first, second and fifth convolutional layer) and three fully connected layers; after each layer ReLu activation is performed except for the last one where instead there is a softmax layer that serves as the classification layer of the network.23,24
Transfer learning is the process of taking a pre-trained network and use it as a starting point to learn a new task. Fine-tuning a network with transfer learning is usually much faster and easier than training a network with randomly initialized weights from scratch. Then, a pre-trained CNN quickly transfer learned features to a new task using a smaller number of training images. In this paper, we applied a transfer learning method to retrain the MatLab - AlexNet CNN for an effective glaucoma and diabetic retinopathy detection to make the testing and procedure accessible (Figure 1).
To carry out the detection of glaucoma and DR through CNN, image pre-processing and processing techniques are required. The different steps are summarized in Figure 2.
For the training of the CNN it is necessary to use retinal fundus images of the eye. Several public databases that compile different eye conditions are available on the internet. In this sense, it is possible to find free access databases and databases with access upon request. The following were used in this work:
- Free access - databases
○ Asia Pacific Tele-Ophthalmology Society (APTOS). Contains 3662 images of diabetic retinopathy that were used in the APTOPS 2019 blindness screening competitions. Each image has been resized and cropped to have a maximum size of 1024px. A certified clinician rated each image according to the severity of diabetic retinopathy on a scale of 0 to 4. A directory file is provided according to the previous scale: No diabetic retinopathy (0), Mild (1), Moderate (2), Severe (3), and Proliferative diabetic retinopathy (4).25
○ High-Resolution Fundus (HRF) Image Database. Contains 15 images of healthy patients, 15 images of patients with diabetic retinopathy and 15 images of glaucomatous patients. They were captured by a Canon CR-1 fundus camera with a field of view of 45 degrees with a resolution of 3504×2336px.26
○ Sungjoon Choi High-Resolution Fundus (sjchoi86-HRF). Created by Sungjoon Choi, assistant professor at Korea University, contains 601 fundus images of different pixel sizes divided into 4 groups: normal (300 images), glaucoma (101 images), cataract (100 images) and retina disease (100 images).27
- Access upon request – databases
○ Large-scale attention based glaucoma (LAG). Contains fundus images with positive (1711 images) and negative glaucoma (3143 images) samples obtained from Beijing Tongren Hospital with a resolution of 500×500px. Each fundus image is diagnosed by qualified glaucoma specialists, taking into consideration of both morphologic and functional analysis.28
○ Ocular Disease Intelligent Recognition (ODIR). Contains images of 5000 patients with various eye diseases collected by Shanggong Medical Technology Co., Ltd. from different hospitals/medical centers in China. The fundus images are captured with various cameras on the market, resulting in varied image resolutions. They classify patients into eight labels based on the images of both eyes. A directory file is provided according to the following label: Normal Fundus (N), Diabetes (D), Glaucoma (G), Cataract (C), Age related Macular Degeneration (A), Hypertension (H), Pathological Myopia (M), Other diseases/abnormalities (O).29
In the case of the ODIR database, photographs labeled in their directory file as “glaucoma” (G) and “normal fundus” (N) were extracted for a total of 200 images and 2873 images, respectively. On the other hand, for the APTOS database, photographs labeled in their directory as “moderate” (2), “severe” (3) and “proliferative diabetic retinopathy” (4) were extracted for a total of 1487 images in general.
The AlexNet architecture design only supports color images (RGB), with a resolution of 227×227px. Convertidor_227_final.m code provides a small user interface that contains three functions (see the Software availability links for deeper descriptions): add a red color filter to grayscale images,30 crop black areas from images, and resize images of any dimension to a new 227×227px image.
The function of cropping black areas in the photograph by Converter_227_final.m is applied to each database. This is done to have more information on the retinal area and eliminate areas of no interest. This function binarizes the original image to obtain a black and white image of equal dimensions. Since the area where a color pixel existed now has a value of 1 and the black areas have a value of 0, the pixel location index by row and column where the value is equal to 1 is extracted as a list. Using the value of the pixel location index as image coordinates, the maximum and minimum value per row and column is determined to establish the cropping edges of the image. It should be mentioned that due to its code design, this function does not affect previously cropped images that no longer contain black areas.
Once the black borders are removed, the function to change the size of the photographs by Converter_227_final.m is applied. Then, all the database images are converted in a single dimension of 227×227px. Since, all databases satisfy the color image format (RGB). It was not necessary to add the red color filter by Converter_227_final.m code. According to their original medical classification, the obtained retinal fundus images were labeled as non-disease (Non_D), suspicious glaucoma (Sus_G) and suspicious diabetic retinopathy (Sus_R). Five storage folders were prepared for the retraining of the CNN (Table 2).
In order to develop predictive software for glaucoma disease, we are going to make use of transfer learning in order to retrain CNN AlexNet. For this, we load the pre-trained AlexNet network and also the different databases (LAG, APTOS, HFR, ODIR, and sjchoi86-HRF) containing the images of the different pathologies to be classified, thus being the glaucoma and retinopathy, besides that we use the information from Refs. 31–38 to develop our algorithm.
In order to begin training with this new dataset, the new images should be unzipped and loaded as ImageData format. AlexNet will automatically label the images based on folder names, corresponding to non-disease fundus eyes images, glaucoma fundus eyes images, and retinopathy fundus eyes images. Then, the model stores the data as an ImageData object, we must divide the data into training and validation data sets. Once the first classification is obtained, the model further divides the data into training images having an equivalent of 70% of the images and the other 30% for validation, for this we use “. splitEachLabel” which splits the images data store into two new data stores.38
Training algorithm for transfer learning
Input ->Retinal fundus images (X, Y); Y = {y {Non-disease, Suspicious-Glaucoma, Suspicious-Diabetic-Retinopathy}
Output-> Re-trained model that classifies the retinal fundus images into respective Y
------------------------------------------------------------------------------------------------------------------
Import the pre-trained model AlexNet Network with its corresponding weights.
Replace the last three layers of the Network:
-Fully connected layer (Set the 'WeightLearnRateFactor' to 20 and the 'BiasLearnRateFactor' to 20; and set its output to the number of elements of Y).
-Softmax layer
-Classification layer
Training-progress settings
MinibatchSize->It is the number of elements into the group of inputs for each iteration
MaxEpoch->It is the maximum number of times that the network is going to use all the input elements
InitialLearnRate ->The learning rate is a tuning parameter that determines the step size at each iteration while moving toward a minimum of a loss function.
Shuffle->It is the action of mixing randomly various elements from our databases
ValidationData ->It is a group of images from the dataset that the network is using to Validate how good the network is getting at classification
ValidationFrequency ->It is the number of iterations that the system does before doing a validation process to assess in real time how the training is going
Verbose->Verbose mode is an option that provides additional details as to what the computer is doing and what drivers and software it is loading during startup
Training Code
options = trainingOptions('sgdm', …
'MiniBatchSize',10, …
'MaxEpochs',6, …
'InitialLearnRate',1e-4, …
'Shuffle','every-epoch', …
'ValidationData',augimdsValidation, …
'ValidationFrequency',3, …
'Verbose',false, …
'Plots','training-progress');
The aforementioned process was applied several times with the different databases acquired. Therefore, we were able to create five new networks based on the structure and weights of the original Alexnet network, the five new networks were denominated: netTransfer 1 (Storage folder 1), netTransfer 2 (Storage folder 2), netTransfer 3 (Storage folder 3), netTranfer 4 (Storage folder 4), and netTransfer 5 (Storage folder 5). The following figure resumes the architecture of all the new networks formed during the transfer learning method (Figure 3).
The results we obtained with our transfer learning code and our different databases over the course of the project was the development of five newly retrained AlexNet networks, to which we will now refer as NetTransfer networks. The confusion matrixes (CM) for the respective NetTransfer networks can be seen in Figure 4 including precision, recall, false positive (FP), false negative (FN) and accuracy values (yellow box). Additionally, the rows represent the known values and columns the predicted values.
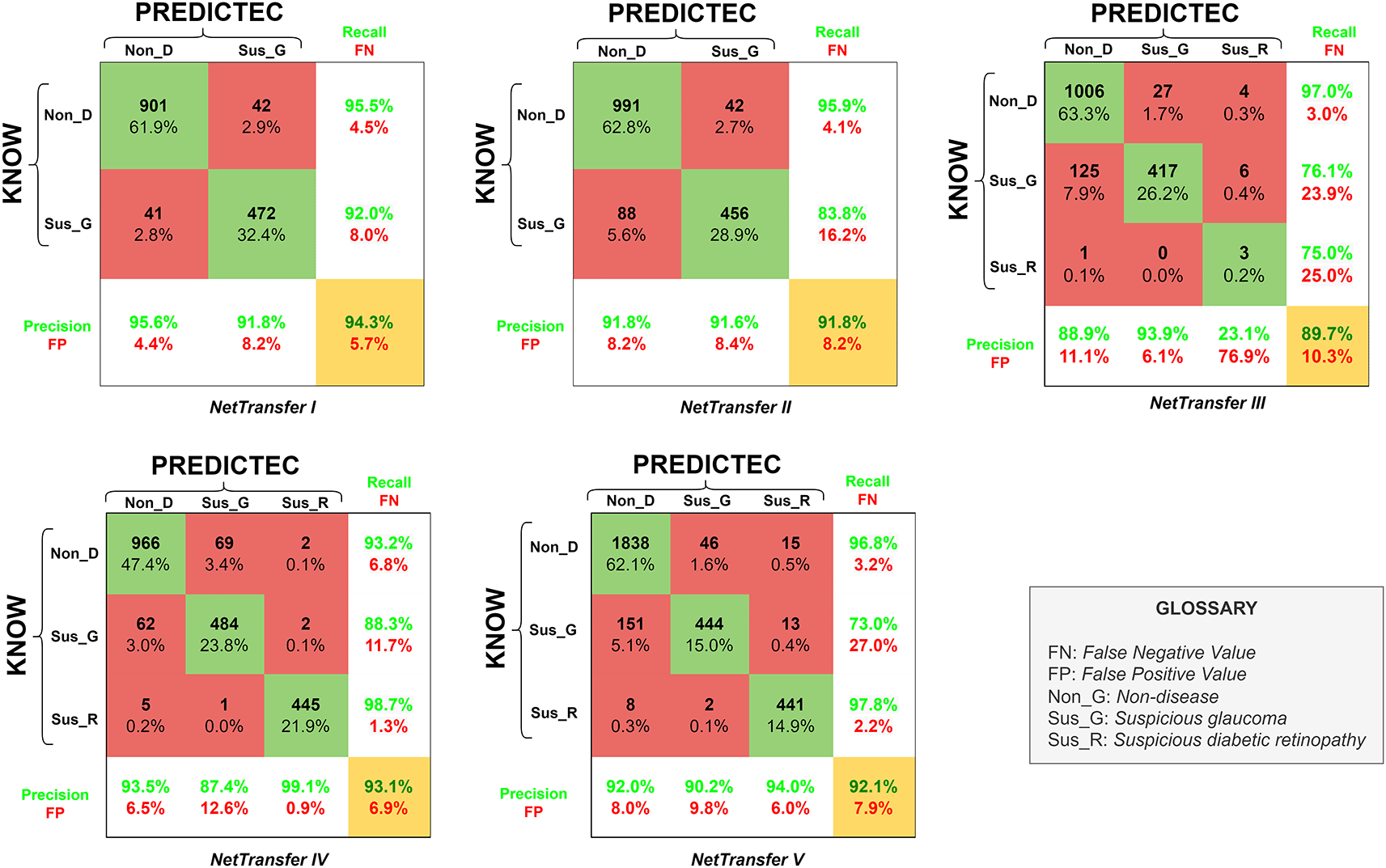
NetTransfer I network was only based on glaucoma and non-disease image cases existing in the LAG-database (Table 2), training with these datasets lead to values of validation accuracy of 94.3%. Besides that, Non_D detection also presented values of 95.5% for recall (4.5% for FN), and values of 95.6% for the precision of the system (4.4% for FP).
NetTransfer II network was based on glaucoma and non-disease images cases existing in the LAG-database and the sjchoi86-HRF database (Table 2), training with these datasets lead to values of validation accuracy of 91.8%. Besides that, Non_D detection presented values of 95.9% for recall (4.1% for FN), and values of 91.8% for the precision of the system (8.2% for FP).
NetTransfer III network was based on glaucoma, diabetic retinopathy and non-disease images cases existing in the LAG-database, sjchoi86-HRF database and the HRF database (Table 2), training with these datasets lead to values of validation accuracy of 89.7%. Besides that, Non_D detection presented values of 97.0% for recall (3.0% for FN), and values of 88.9% for the precision of the system (11.1% for FP).
NetTarnsfer IV network was based on glaucoma, diabetic retinopathy and non-disease images cases existing in the LAG-database, sjchoi86-HRF database, HRF database and the APTOS database (Table 2), training with these datasets lead to values of validation accuracy of 93.1%. Besides that, Non_D detection presented values of 93.2% for recall (6.8% for FN), and values of 93.5% for the precision of the system (6.5% for FP).
NetTransfer V network was based on glaucoma, diabetic retinopathy and non-disease images cases existing in the LAG-database, sjchoi86-HRF database, HRF database, APTOS database and ODIR database (Table 2), training with these datasets lead to values of validation accuracy of 92.1%. Besides that, Non_D detection presented values of 96.8% for recall (3.2% for FN), and values of 92.0% for the precision of the system (8.0% for FP).
Several works were presented for glaucoma detection using fundus photographs by calculating cup disk ratio (CDR). For example, Carrillo and coworkers39 developed an autonomic detection method and a novel method for cup segmentation with a percentage of success of 88.5% (Table 3). Another work from Anum Abdul and peers,40 an algorithm was provided to detect CDR and hybrid textural and intensity features. Those features were used to classify the autonomous system, and it gave improvements in the results from previous studies that only used CDR, thanks to their hybrid approach, they reached an accuracy of 92% (Table 3). Even though we did not make use of CDR characteristic, our AlexNet approach seems to reach comparable levels of accuracy (92.1%) to these previously mentioned methods without the need to calculate the CDR.
| Detectable pathology | Detection method | Dataset (Size) | Type of channels | Number of classes | Performance measure | Ref. | |||
|---|---|---|---|---|---|---|---|---|---|
| Accuracy (%) | Sensitivity (%) | Specificity (%) | AUC | ||||||
| Glaucoma and diabetic retinopathy (Ours) | CNN | LAG, APTOS, HRF, and ODIR (9860) | RGB | 3 | 92.06 | 73.0 (Glaucoma) 97.80 (Retinopathy) | 97.93 (Glaucoma) 98.78 (Retinopathy) | - | - |
| Glaucoma | Algorithm to improve glaucoma detection using cup segmentation | Set of fundus images from the CPAG in Bucaramanga, Colombia | Gray scale, each RGB color channel independently | 2 | 88.50 | - | - | - | 39 |
| Glaucoma | Algorithm to detect glaucoma using a fusion of CDR and hybrid textural and intensity features | Local database of 50 fundus images with 15 glaucoma and 35 healthy images | Binary image, Green and RGB | 2 | 92.00 | 100.00 | 88.00 | - | 40 |
| Glaucoma | DCNN | ORIGA (650) and SCES (1676) | RGB | 2 | - | - | - | 0.83 (ORIGA) and 0.88 (SCES) | 41 |
| Glaucoma | CNN | CGSA (241032) | Gray scale and RGB | 3 | - | 82.20 | 70.40 | 0.82 | 22 |
| Glaucoma | Direct Feed Neural Network | ACRIMA (705) | Green, gray scale and binary image | 2 | 94.61 | 94.57 | 95.00 | - | 42 |
| Diabetic retinopathy | Customized DCNN | EyePacs, MESSIDOR 2 and E-Ophtha (for testing) (75137) | RGB | 2 | - | 74.00-94.00 | 80.00-98.00 | 0.94-0.97 | 17 |
| Diabetic retinopathy | CNN (modified AlexNet) | Messidor (1190) | Green and RGB | 4 | 95.60-96.60 | 88.00-96.00 | 97.30-96.60 | - | 18 |
| Diabetic retinopathy | GoogleNet | Kaggle (200) | RGB | 5 | 88.00 | 75.00 | 52.00 | - | 16 |
| Diabetic retinopathy | Fused CNN512, CNN299, and CNN (YOLOv3, EfficientNetB0) | DDR (13673), and Kaggle (3662) | RGB | 5 | 89.00 | 89.00 | 97.30 | 0.97 | 15 |
In other more rigorous studies such as Xiangyu Chen work,41 a deep CNN was developed with a total of six layers: two fully connected layers and four convolutional layers. The results drop scores of prediction from 71% to 83% from real images (Table 3). On the other hand, Hanruo Liu and peers22 made a deep learning system using a total of 241,032 images from 68,013 patients. In this work, every image was subjected to a multiple layers of grading system, in which graders were from students to senior specialists on glaucoma, from these they obtained good levels of sensitivity and specificity (82.20% and 70.40%). When compared to these other CNN systems, our AlexNet based detection system still presented comparable and even arguably better results in accuracy, sensitivity and specificity for glaucoma and DR detection (Table 3).
Another related work from Almeida and peers,42 uses image processing in MATLAB to improve the accuracy of glaucoma tests by extracting the most pertinent qualities of the images obtaining promising results with an accuracy, specificity, and sensitivity greater than 90% (Table 3), which indicates that it gives an excellent start for us to assess the glaucoma diagnosis through AI. While Almeida and co-workers seems to be better suited for glaucoma detection, our AlexNet system presented the advantage of also being able to detect more pathologies at the same time, such as DR, which was the other disease we decided to add to our detection system.
As our system is also able to detect DR, it is also pertinent to compare it to other deep learning systems that were developed for DR detection. In the study realized by Rishab Gargeya and Theodore Leng,17 they developed and evaluated a data-driven deep learning algorithm as a novel diagnostic tool for automated DR detection, which proved to reach high efficacy computer-aided model, with low-cost, which lead to correct DR diagnostics without depending on clinicians to examine and grade images manually (Table 3). A different study made by Shanthi and Sabeenian,18 used a modified AlexNet CNN system for the detection of DR in a big data training of the network (Table 3). Additionally, Amnia Salma and peers16 develop a similar system, but the use GoogleNet instead of AlexNet (Table 3). While all of these systems follow similar principles to our propose system, it is important to remark that our AlexNet pretrained network acquired higher accuracies, sensitivities and specificities than the previously mentioned systems, mostly due to using a higher number datasets. Also we decided to try to go beyond and used AlexNet to classify more diseases at the same time. The following table summarizes and compares the capabilities of detection of our AlexNet pathology detection system to all the previously mentioned works, as well as some other important studies that were not addressed (Table 3).
Additionally, for the implementation of the AlexNet architecture on open-source language, we endorse the use of TensorFlow which is a free open-source self-learning platform based on the Python language, mainly developed by Google.43 Among its many libraries we can find Keras, which is a deep learning application programming interface (API) developed for Python built on TensorFlow, from which we can build our AlexNet equivalent model. The recommended model is the sequential model of Keras which allows a user to define the model as a series of convolutional layers with max pooling.44
In the presented research, the training of a CNN through the use of MATLAB software and its AlexNet tool, allowed the effective recognition of two eye diseases (glaucoma and DR) through retinal fundus images. Additionally, the use of open access databases allows the replicability and reproducibility of the present study. Being the APTOS, HRF and sjchoi86-HRF databases of immediate access. Meanwhile, LAG and ODIR are databases with access upon request. The implementation of the different databases (LAG, APTOS, HRF, ODIR, sjchoi86-HRF), proved to be effective in improving the prediction percentages of the different neural network trainings.
In general, the most common eye affections are presented through a series of symptoms, such as blurred vision, spots, glare, eye fatigue, dry eyes, among others. In this way, glaucoma proves to be a condition that damages the optic nerve and generally does not present any symptoms, until the person suffering from it perceives a decrease in vision in the final stages of the disease. Based on the foregoing, it is necessary to create tools that allow an effective detection of this type of affectation, for example CNN systems as an alternative, highly reliable in the automation of processes. Additionally, this research moved its detection objective to the addition of a new disease, such as DR.
Future improvements to this algorithm could include the creation of a more user-friendly graphical interface for users who are not experts in programming language. In this way, the detection tasks will be based on the selection of options and not on the coding of algorithms. On the other hand, as previously mentioned, it is possible to replicate the AlexNet-CNN using Python, by using existing tools such as TensorFlow and Keras API. Therefore, a subsequent study will concentrate efforts on implementing the recognition system in the open-source language, to endorse the use of non-proprietary software in order to increase reproducibility.
MatLab codes and scripts related to image processing, pre-processing & Training versions of the AlexNet Convolutional Neural Network (NetTransfers I-V).
Source code available from: https://github.com/IscArias/EyeEvaluationSourceCode
Archived source code as at time of publication: https://doi.org/10.5281/zenodo.709887945
License: 2-Clause BSD License
Free access - databases:
- Asia Pacific Tele-Ophthalmology Society (APTOS). The data download process is free and does not require authorization or registration. Follow the directions of the data host to reference the database. Link: https://www.kaggle.com/c7934597/resized-2015-2019-diabetic-retinopathy-detection/metadata/
- High-Resolution Fundus (HRF) Image Database. The data download process is free and does not require authorization or registration. Follow the directions of the data host to reference the database. Link: https://www5.cs.fau.de/research/data/fundus-images/
- Sungjoon Choi High-Resolution Fundus (sjchoi86-HRF). The data download process is free and does not require authorization or registration. Follow the directions of the data host to reference the database. Link: https://github.com/cvblab/retina_dataset
Access upon request – databases:
- Large-scale attention based glaucoma (LAG). Database for academic purposes. An email must be written to the hosts who will provide a password to authorize the download. Once the key is obtained and entered, the download is free. It is not necessary to register on any additional platform. Follow the directions of the data host to reference the database. Link: https://github.com/smilell/AG-CNN
- Ocular Disease Intelligent Recognition (ODIR). Database for academic purposes. It is required to create an account and register on the platform, providing data on the user's institution, department and country. Once registered, a request must be submitted in order to download the database. Authorized access, the user can download the database freely. Follow the directions of the data host to reference the database. Link: https://odir2019.grand-challenge.org/dataset/
Additional Codes for Image Evaluation, Image Selection from Data Bases and Confusion Matrix Plotting.
Source code available from: https://github.com/IscArias/EyeEvaluationSourceCode_Extra
Archived source code as at time of publication: https://doi.org/10.5281/zenodo.710261846
License: 2-Clause BSD License
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Partly
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Ophthalmology; deep learning; machine learning; fundus photography
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
Partly
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: machine learning, computer vision, eye diseases, biometric identification.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 2 (revision) 03 Apr 24 |
read | read |
|
Version 1 05 Jan 23 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)